Abstract
Background
The exploration of genetic diversity is the key source of germplasm conservation and potential to broaden its genetic base. The globally growing demand for chickpea suggests superior/climate-resilient varieties, which in turn necessitates the germplasm characterization to unravel underlying genetic variation.
Methodology and results
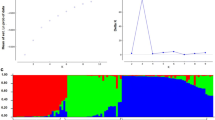
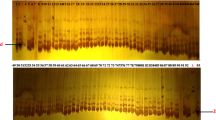
A chickpea core collection comprising of diverse 192 accessions which include cultivated Cicer arietinum, and wild C. reticulatum, C. echinospermum, and C. microphyllum species were investigated to analyze their genetic diversity and relationship, by assaying 33 unlinked simple sequence repeat (SSR) markers. The results amplified a total of 323 alleles (Na), ranging from 2 to 8 with an average of 4.25 alleles per locus. Expected heterozygosity (He) differed from 0.46 to 0.86 with an average of 0.68. Polymorphic information content (PIC) ranged from 0.73 to 0.98 with an average of 0.89. Analysis of molecular variance (AMOVA) showed that most of the variation was among individuals (87%). Cluster analysis resulted in the formation of four distinct clusters. Cluster I represented all cultivated and clusters II, III, and IV comprised a heterogeneous group of cultivated and wild chickpea accessions.
Conclusion
We report considerable diversity and greater resolving power of SSR markers for assessing variability and interrelationship among the chickpea accessions. The chickpea core is expected to be an efficient resource for breeders for broadening the chickpea genetic base and could be useful for selective breeding of desirable traits and in the identification of target genes for genomics-assisted breeding.



Similar content being viewed by others
Data availability
The data analyzed and generated during the current study are available within the article [and/or] its supplementary materials.
References
Mallikarjuna BP, Samineni S, Thudi M, Sajja SB, Khan AW et al (2017) Molecular mapping of flowering time major genes and QTLs in chickpea (Cicer arietinum L.). Front Plant Sci 8:1140
Archak S, Tyagi RK, Harer PN, Mahase LB, Singh N, Dahiya OP et al (2016) Characterization of chickpea germplasm conserved in the Indian national genebank and development of a core set using qualitative and quantitative trait data. Crop J 4(5):417–424
FAOSTAT, Food and Agriculture Organization of the United Nation Statistics for (2017) Online database at https://www.fao.org/faostat/en/#data
Annual Report DPD. 2017. Directorate of Pulses Development, Ministry of Agriculture and Farmers Welfare, Government of India. DPD/Pub./TR/29/2017–18
Varshney RK, Song C, Saxena RK, Azam S, Yu S, Sharpe AG et al (2013) Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat Biotechnol 31(3):240
Jain M, Misra G, Patel RK, Priya P, Jhanwar S, Khan AW (2013) A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.). Plant J 74:715–729
Upadhyaya HD, Yadav D, Dronavalli N, Gowda CLL, Singh S (2010) Mini core germplasm collections for infusing genetic diversity in plant breeding programs. Electron J Plant Breed 1(4):1294–1309
Nguyen TT, Taylor PWJ, Redden RJ, Ford R (2004) Genetic diversity estimates in Cicer using AFLP analysis. Plant Breed 123(2):173–179
Sethy NK, Shokeen B, Edwards KJ, Bhatia S (2006) Development of microsatellite markers and analysis of intra specific genetic variability in chickpea (Cicer arietinum L.). Theor Appl Genet 1(12):1416–1428
Fayaz H, Mir AH, Tyagi S, Wani AA, Jan N, Yasin M, Mir JI, Mondal B, Khan MA, Mir RR (2021) Assessment of molecular genetic diversity of 384 chickpea genotypes and development of core set of 192 genotypes for chickpea improvement programs. Genet Resour Crop Evol. https://doi.org/10.1007/s10722-021-01296-0
Talebi REZA, Fayaz F, Mardi M, Pirsyedi SM, Naji AM (2008) Genetic relationships among chickpea (Cicer arietinum) elite lines based on RAPD and agronomic markers. Int J Agric Biol 10(3):301–305
Jida Z, Alemu A, Mullualem D (2018) Genetic diversity among elite chickpea (Cicer arietinum L.) varieties of ethiopia based on inter simple sequence repeat markers. Afr J Biotechnol 17(34):1067–1075
Sefera T, Abebie B, Gaur PM, Assefa K, Varshney RK (2011) Characterisation and genetic diversity analysis of selected chickpea cultivars of nine countries using simple sequence repeat (SSR) markers. Crop Pasture Sci 62(2):177–187
Keneni G, Bekele E, Imtiaz M, Dagne K, Getu E, Assefa F (2012) Genetic diversity and population structure of ethiopian chickpea (Cicer arietinum L.) germplasm accessions from different geographical origins as revealed by microsatellite markers. Plant Mol Biol Rep 30(3):654–665
Ghaffari P, Talebi R, Keshavarzi F (2014) Genetic diversity and geographical differentiation of Iranian landrace, cultivars, and exotic chickpea lines as revealed by morphological and microsatellite markers. Physiol Mol Biol Plants 20(2):225–233
Aggarwal H, Choudhary SP, Rana MK, Choudhary R (2018) Assessment or evaluation of genetic diversity among 66 cultivars of chickpea (Cicer arietinum L.) of Indian origin Using SSR MARKERS. Int J Curr Microbiol Appl Sci 7(2):523–533
Choudhary P, Khanna SM, Jain PK (2012) Genetic structure and diversity analysis of the primary gene pool of chickpea using SSR markers. Genet Mol Res 11(2):891–905
Mir AH, Bhat MA, Dar SA, Sofi PA, Bhat NA, Mir RR (2021) Assessment of cold tolerance in chickpea (Cicer spp.) grown under cold/freezing weather conditions of North Western Himalayas of Jammu and Kashmir, India. Physiol Mol Biol Plants. https://doi.org/10.1007/s12298-021-00997-1
Fayaz H, Rather IA, Wani AA, Tyagi S, Pandey R, Mir RR (2019) Characterization of chickpea gene pools for nutrient concentrations under agro-climatic conditions of North- Western Himalayas. Plant Genet Resour 17:464–467
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer length polymorphism in barley: mendelian inheritance, chromosomal location and population dynamic. Proc Natl Acad Sci USA 81:8014–8019
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci, USA 70:3321–3323
Perrier X, Jacquemoud-Collet JP (2006) Darwin software. http://darwin.cirad.fr/darwin.
Singh R, Sharma P, Varshney RK, Sharma SK (2008) Chickpea improvement: role of wild species and genetic markers. Biotechnol Genet Eng 25:267–313
Maesen LJG (1972) Cicer L, A Monograph of the Genus, with Special Reference to the Chickpea (Cicer arietinum L). Its Ecology and Distribution (Doctoral thesis) Wageningen University
Upadhyaya HD, Oritz R (2001) A mini core subset for capturing diversity and promoting utilization of chickpea genetic resources in crop improvement. Theor Appl Genet 102:1292–1298
Mir RR, Varshney RK (2013) Future prospects of molecular markers in plants. In: Henry RJ (ed) Molecular markers in plants. Blackwell Publishing Ltd, Oxford, pp 170–190
Mir RR, Hiremath PJ, Riera-Lizarazu O, Varshney RK (2013) Evolving molecular marker technologies in plants: From RFLP to GBS. In: Lubberstedt T, Varshney RK (eds) Diagnostics in plant breeding. Springer, Dordrecht, pp 229–250
Saxena MS, Bajaj D, Kujur A, Das S, Badoni S, Kumar V et al (2014) Natural allelic diversity, genetic structure and linkage disequilibrium pattern in wild chickpea. PLoS ONE 9(9):e107484
Upadhyaya HD, Dwivedi SL, Baum M, Varshney RK, Udupa SM, Gowda CLL (2008) Genetic structure, diversity, and allelic richness in composite collection and reference set in chickpea (Cicer arietinum L.). BMC Plant Biol 8:106
Imtiaz M, Materne M, Hobson K, Van Ginkel M, Malhotra RS (2008) Molecular genetic diversity and linked resistance to ascochyta blight in Australian chickpea breeding materials and their wild relatives. Aust J Agric Res 59(6):554–560
Castro P, Millan T, Gil J, Merida J, Garcia ML, Rubio J, Fernadez-Romero MD (2011) Identification of chickpea cultivars by microsatellite markers. J Agric Sci 149(4):451–460
Khan R, Khan H, Harada F, Harada K (2010) Evaluation of microsatellite markers to discriminate induced mutation lines, hybrid lines and cultigens in chickpea (’Cicer arietinum L.). Aust J Crop Sci 4(5):301
Shukla N, Babbar A, Prakash V, Tripathi N, Saini N (2011) Molecular diversity analysis in some promising lines of chickpea using SSR markers. J Food Legumes 24(4):320–323
Samyuktha SM, Kannan JR, Geethanjali S (2018) Molecular genetic diversity and population structure analysis in chickpea (Cicer arietinum L.) germplasm using SSR markers. Int J Curr Microbiol Appl Sci 7(2):639–651
Mohan S, Kalaimagal T (2020) Genetic diversity studies in chickpea (Cicer arietinum L.) genotypes using SSR markers. J Food Legumes 33(1):1–5
Udupa SM, Sharma A, Sharma RP, Pai RA (1993) Narrow genetic variability in Cicer arietinum L. as revealed by RFLP analysis. J Plant Biochem Biotechnol 2(2):83–86
Sonnante G, Marangi A, Venora G, Pignone D (1997) Using RAPD markers to investigate genetic variation in chickpea. J Genet Breed 51:303–307
Sant VJ, Patankar AG, Sarode ND (1999) Potential of DNA markers in detecting divergence and in analyzing heterosis in Indian elite chickpea cultivars. Theor Appl Genet 98:1217–1225
Singh R, Prasad CD, Singhal V, Randhawa GJ (2003) Assessment of genetic diversity in chickpea cultivars using RAPD, AFLP, and STMS markers. J Breed Genet 57:165–174
Hajibarat Z, Saidi A, Hajibarat Z, Talebi R (2014) Genetic diversity and population structure analysis of landrace and improved chickpea (Cicer arietinum) genotypes using morphological and microsatellite markers. Environ Exp Biol 12:161–166
Saeed A, Hovsepyan H, Darvishzadeh R, Imtiaz M, Panguluri SK, Nazaryan R (2011) Genetic diversity of Iranian accessions, improved lines of chickpea (Cicer arietinum L.) and their wild relatives by using simple sequence repeats. Plant Mol Biol Rep 29(4):848–858
Khamassi K, Bettaiëb Ben Kaab L, Khoufi S, Chaabane R et al (2012) Morphological and molecular diversity of Tunisian chickpea. Eur J Hortic Sci 77(1):31
Bharadwaj C, Chauhan SK, Yadav S, Tara Satyavathi C, Singh R, Kumar J, Srivastava R, Rajguru G (2011) Molecular marker-based linkage map of chickpea (Cicer arietinum) developed from desi × kabuli cross. Int J Agric Sci 81(2):116–118
Sachdeva S, Bharadwaj C, Sharma V, Patil BS, Soren KR, Roorkiwal M, Varshney RK, Bhat KV (2018) Molecular and phenotypic diversity among chickpea (Cicer arietinum L.) genotypes as a function of drought tolerance. Crop Pasture Sci 69(2):142–153
Excoffier L, Smouse PE, Quattro J (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131(2):479–491
Roorkiwal M, Von Wettberg EJ, Upadhyaya HD, Warschefsky E, Rathore A, Varshney RK (2014) Exploring germplasm diversity to understand the domestication process in Cicer spp using SNP and DArT markers. PLoS ONE 9(7):e102016
Van Oss R, Abbo S, Eshed R, Sherman A, Coyne CJ et al (2015) Genetic relationship in Cicer sp expose evidence for geneflow between the cultigen and its wild progenitor. PLoS ONE 10(10):e0139789
Parween S, Nawaz K, Roy R, Pole AK, Venkata S, Misra G, Jain M et al (2015) An advanced draft genome assembly of a desi type chickpea (C. arietinum L.). Sci Rep 5:12806
Thudi M, Upadhyaya HD, Rathore A, Gaur PM et al (2014) Genetic dissection of drought and heat tolerance in chickpea through genome-wide and candidate gene-based association mapping approaches. PLoS ONE 9(5):e96758
Bharadwaj C, Srivastava R, Chauhan SK, Satyavathi CT, Kumar J, Faruqui A et al (2011) Molecular diversity and phylogeny in geographical collection of chickpea (Cicer sp.) accessions. J Genet 1:7
Acknowledgements
The authors are thankful to ICRISAT (Hyderabad), IIPR (Kanpur), NBPGR (New Delhi) for providing the germplasm for the present study.
Funding
This study was not funded by any external funding agency.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies/ research with human or animal subjects.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mir, A.H., Bhat, M.A., Fayaz, H. et al. SSR markers in revealing extent of genetic diversity and phylogenetic relationships among chickpea core collection accessions for Western Himalayas. Mol Biol Rep 49, 11469–11479 (2022). https://doi.org/10.1007/s11033-022-07858-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07858-4