Abstract
Background
The Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-2) has had a major impact on world health over the last 2 years. The emergence of SARS-CoV-2 variants, particularly concerning variants, may affect the virus’s pathogenicity, transmissibility, and vaccines potency. Both delta and the omicron variants have been designated by WHO as variants of concern.
Methods and results
In this study, molecular techniques such as qPCR, conventional PCR, and sequencing were used to identify the first SARS-CoV-2 omicron variant that circulated in Iraq in January 2022. Bioinformatics and computational tools like phylogenetic analysis, predicted physical and chemical properties, stability, and molecular docking of the spike protein were used to compare the omicron with the delta variants. We found the receptor binding domain (RBD) and spike protein in omicron contain a greater number of hydrophobic amino acids compared to delta variant. We discovered a disorder–order conversion in RBD regions of the omicron variant, and this change may be important in terms of the effect of disordered residues/regions on spike protein stability and interaction with human angiotensin converting enzyme 2 (ACE2). Docking studies show that the omicron variant requires less energy to engage with ACE2, contributing to its higher binding affinity with human ACE2, consistent with more contagious transmission.
Conclusion
This is the first molecular study of the circulated omicron and delta variants in Iraq, showing that the omicron variant in Iraq had a higher affinity for ACE2 than the delta variant, which may lead to higher transmissibility.
Similar content being viewed by others
Introduction
The Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-2) caused COVID-19 pandemic has had a major impact on world health and the economy [1]. Over the last 2 years, the world has been on high COVID-19 alert subsequent to the SARS-CoV-2 outbreak, which has killed more than 5 million people. New SARS-CoV-2 mutations have been reported on a weekly, if not daily, basis, and it continues to mutate and take on new forms as long as it is spread over the world [2, 3]. Therefore, numerous vaccines have been created and used to limit the spread of the SARS-CoV-2 virus [4]. However, these efforts are not quite restricted to emerging new variants [5].
On November 11, 2021, and November 14, 2021, samples from Botswana and South Africa showed a large number of mutations [6]. On November 26, 2021, these variants were classified as a virus of concern (VOC) by the WHO. They were given a new name, omicron variant (B.1.1.529). The omicron variant has an accumulation of a large number of non-synonymous mutations, notably in the spike protein, which is responsible for the commencement of infection by means of cell entrance. There are 15 mutations in the spike protein’s receptor binding domain (RBD), while the entire S gene for that spike protein has 32 non-synonymous mutations [7]. Viruses recognize human ACE2 receptors through their RBD, and once the RBD-ACE2 complex is formed, the virus attaches to human cells, where it inserts into the cell and causes infection [8].
In Iraq, SARS CoV-2 was managed and monetarized after the first identification of the disease [9]. In the second half of 2020, B.1.428.1 looked to be the dominant lineage, but in early 2021, the alpha strain (B.1.1.7) looked to be the dominant lineage [10].
The objective of the present study is to perform molecular characterization of the newly emerging omicron variant in Iraq and to examine the binding affinity of omicron and delta variants to ACE2, utilizing a number of computational techniques. This research is supposed to shed some light on how the virus has changed and the ability to spread in the area.
Methodology
Sample collection
In this study, two asymptomatic SARS CoV-2 carriers were screened. Both were males, the ages of 35 and 39 years old. The nasopharyngeal swab samples were collected and stored in viral transport media (VTM) at − 70 °C in the COVID-19 Molecular Diagnostic Laboratory of Shahid Tahir Hospital in Sulaimani, Kurdistan Region of Iraq.
Amplification of S gene
In biosafety laboratory level 2, the samples were extracted by the AddPrep Viral Nucleic Acid Extraction Kit (Addbio, South Korea). Direct reverse transcription and S gene amplification were performed on the isolated nucleic acid using AddScript one step RT-PCR Master mix (Addbio, South Korea). The reaction was done in 0.2 mL PCR tubes containing 10 µL master mix, 4 µL DEPC water, 5 µL RNA sample, and 1 µL (10 pmol) of each primer (forward and reverse) (Table S1).
The Techne TC-4000 thermal cycler (Techne Ltd., UK) was set for 30 min at 50 °C, 10 min of denaturation at 95 °C. The sample was then subjected to 40 cycles of denaturation (95 °C for 30 s), annealing (55 °C for 80 s), and extension (72 °C for 50 s). Then, 5 min at 72 °C was set as the last extension.
Loading 6 µL of PCR product onto a 1% agarose gel in TBE solution was used to evaluate the PCR results. 8 µL of safe gel stain dye were used to color the gel (Addbio, South Korea). The electrophoresis unit was operated at 120 V for 40 min. The PCR amplicons were analyzed according to 100 bp DNA marker migration.
Design of primer sets for genome sequencing
In order to get nearly complete genome sequences of the SARS-CoV-2 spike glycoprotein, we used the Primer 3 plus and the NCBI Primer Blast web-based primer design tools to make four primer pairs. These primers are capable of amplifying all SARS-CoV-2 variations, including the omicron. All of the primers were manufactured by Macrogen® in South Korea (Table S1).
Sequencing and phylogenetic of the S gene
Purification of the PCR product was performed using the AddPrep PCR Purification Kit according to the manufacturer's instructions (Addbio, South Korea). They were then sequenced using the Sanger method at a sequencing facility in South Korea. The identification of each nucleotide sequence was validated using reverse and forward primer sequencing reactions.
After obtaining the sequences, they were assembled using NCBI alignments. The appropriate reading frames of the viral sequences included in the spike glycoprotein were determined using the ExPASy translate program [11]. The coding sequences were deposited in the global initiative on sharing avian flu data (GISAID). The viruses were given a name and allocated an accession ID, (hCoV-19/Iraq/Sulaimani-8/2021) EPI_ISL_9247320, for the SARS-CoV-2 delta variant, and (hCoV-19/Iraq/Sulaimani-1/2022) EPI_ISL_9246344, for the SARS-CoV-2 omicron variant. The spike gene sequences were used to construct a phylogenic tree with different virus variants in the world (Fig. 1). Nextclade was used to perform phylogenetic analysis based on the GISAID clades [12]. Variants filtering to only those that emerged in 2021 and the wild type, Wuhan-Hu-1, was used as an outgroup to root the trees.
Phylogenetic SARS-CoV-2 variant analysis. The phylogenetic tree was constricted based on sequence date in GISAID data using the Nextstrain server (Nextclade tool). The color indicates the GISAID clades, the red circle indicates the Iraq SARS-CoV-2 variants. The delta variant hCoV-19/Iraq/Sulaimani-8/2021 clusters in (21J delta). The omicron variant, hCoV-19/Iraq/Sulaimani-1/2022, is clustered in 21K omicron. (Color figure online)
Analyzing physical and chemical parameters
The ExPASy ProtParam online tool was used to evaluate the omicron and delta variant amino acid sequences. as well as the theoretical pI and molecular weight. ProtParam also determines the amino acid composition and atomic makeup and products extinction coefficients and half-lives predicted as well as instability indices and aliphatic indices.
Spike protein secondary structure prediction
The secondary structure of the omicron and delta variants was predicted by means of GOR 4. Secondary protein structures may be analyzed using information models and Bayesian statistics using the Garnier–Osguthorpe–Robson (GOR) tool. So, the main objective of GOR is to acquire information in order to develop secondary structure variations.
Determination of conserved and mutated residues
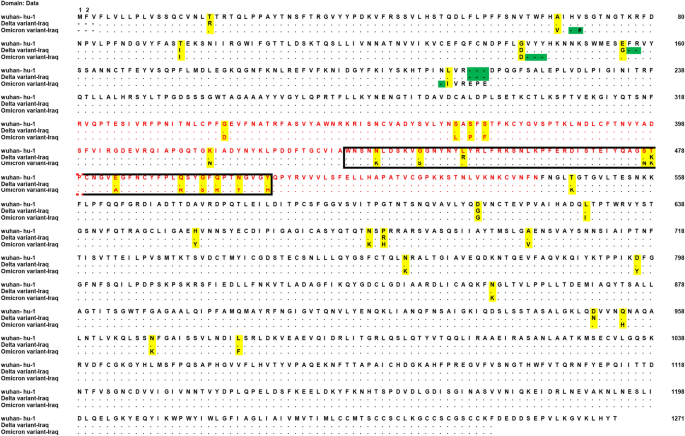
The Wuhan Hu1 (wild type) sequence was aligned with the omicron and delta variant amino acid sequences using the Clustal W algorithm. The alignment figure was created using Mega 10, a bioinformatics software package (Fig. 2).
A sequence alignment of the spike amino acid sequences of SARS-CoV-2 wild type, omicron, and delta variants discovered in Iraq. The yellow highlights indicate non synonymous mutations, and the green highlights indicate indel mutations. The red letters indicate the RBD amino acid, while the box indicates the RBM amino acid. The amino acids are numbered according to the Wuhan-Hu-1 wild type. (Color figure online)
Prediction of modifications to spike protein
In this study, several approaches were used to demonstrate the location of mutations in the spike protein of the variants. We used the SWISS-MODEL web server to convert the amino acid sequence of both the delta and omicron variants into homology models in PDB format [13]: PyMOL v2.4 was then used to look at the three-dimensional structure of the SARS-CoV-2 spike protein [14]. The schematic figures were then edited and merged by using the BioRender platform (https://www.biorender.com). This method was used to clearly show the mutated amino acid in the SARS-CoV-2 surface protein.
Prediction of an intrinsically unstructured protein
Polypeptide segments that do not contain enough hydrophobic amino acids to facilitate co-operative folding are known as intrinsically disordered regions (IDRs). Instead, they are more likely to contain polar or charged amino acids in their composition [15]. The spike protein of omicron variant and delta variant were subjected to prediction using the natural disordered region predictor (PONDR) [16].
Modeling protein stability
The I-Mutant3.0 [17] prediction was used to predict the stability of delta and omicron sequences. Single point mutations may alter protein stability, and this technique uses support vector machines to anticipate such changes. It can also be used to predict the sign of a mutation's effect on protein stability and a regression estimator to predict the related DeltaG.
Prediction of disease-related symptoms
VarSite is a web service that maps known disease-associated variations from Protein Data Bank 3D structures. This website shows how these variants are linked with diseases [18]. Varsite was used to predict SARS-CoV-2 mutations. The data may be used to determine if a mutation is pathogenic or without risk.
Docking of variant RBD and human ACE2
PyMOL used to isolate the RBD sequence monomer (chain A) from the spike trimer protein for both delta and omicron variants, which then used for protein docking. The crystal structure of ACE2 was extracted also by PyMOL from the protein data bank (1R42). To assess the interaction of SARS-CoV-2 variants spike protein with human ACE2 cellular receptors, molecular docking methods from the ClusPro server were utilized in conjunction with the co-crystal structure of the SARS-CoV-2 Spike RBD and human ACE2 cellular receptor contact [19].
Result
Molecular identification and phylogenic analysis of two SARS-CoV-2 in Sulaimani province, Kurdistan region of Iraq, indicated two different variants. The first one belongs to nextclade 21J (delta), GISAID clade O, VOC delta (B.1.617.2.). The second virus belongs to nextclade 21K (Omega), GISAID clade GRA, VOC omicron (B.1.1.529+BA.1) (Fig. 1). According to multiple alignments, the identified omicron variant has 32 spike protein substitution mutations, 3 insertion mutation, 6 deletion mutation as compared with the wild type of Wu-Han-1. About half of these mutations are in the RBD (Fig. 2). While the delta variant spike protein has 9 substitution mutations, and 2 deletion mutations are found in the RBD (Fig. 2).
The physical properties spike protein
Although the delta variant has 1271 amino acids, this study generates only 1251 amino acids, corresponding to the 1250 amino acids of the omicron variant. Due to a deletion mutation, the omicron variants include only one residue less than the delta variant. However, the identified delta variant has a molecular weight of 138,693.76 with a theoretical pI of 7.39, while the identified omicron has a molecular weight of 139,051.56 with a theoretical pI of 7.73. Protein stability of the omicron spike protein showed 34.23 as compared to delta variant 32.45. The aliphatic index of the omicron spike protein is 85.14, as compared to 84.84 in the delta variant. Both SARS-CoV-2 variants share the same percentage of some amino acids in spike protein. However, there is an increase in lysine (K), glutamic acid (E), aspartic acid (D), phenylalanine (F) and isoleucine (I), in the identified omicron variant compared to the delta variant, which is more hydrophobic (Table S2).
Prediction of changes in secondary structural elements
The GOR.4 tool prediction indicates that the omicron variant has more alpha-helix structures (23.84%) but fewer extended strand structures (19.52%) than the delta variant (Table S3). The RBD and RBM of the identified omicron variant have a higher percentage of alpha helices than the delta variant. However, the random coil structure is increased to some extent in RBD and decreased in RBM of the omicron variant (Table S3).
Prediction of an intrinsically unstructured protein
In this study, the inherent disorder of omicron and delta variants was predicted using PONDR® VLXT. The identified omicron variant has a lesser amount of disordered (6.56) region than the circulated delta variant (7.99). In addition, the delta variant RBD showed disordered residues in the amino acid residues E471, I472, and Y473 (Wuhan-Hu-1 numbering). However, the same RBD region of omicron showed no disorder residues.
Protein stability alterations are predicted as a result of mutation
According to the results of an I-Mutant protein stability tool, all amino acid alterations in the delta variate diminish spike protein stability by the same amount. The omicron variant is less stable than the delta variant, with the exception of the N501Y mutation. SIFT analysis showed that the delta variant D950N mutation has affected protein function. Other mutations were shown to be tolerable. A number of non-synonym mutations in the circulated omicron affect protein function, including the N764K, Y505H, and N211I mutations. However, all other mutations are tolerated (Table S4). As determined by the VarSite analysis, mutations with a value greater than 1 have a larger possibility of being disease-causing mutations than variants with values less than or equal to 1 (Table S4).
Discussion
The SARS-CoV-2 caused COVID-19, which is now circulating and causing pandemics around the world. Due to overspreading and multiple mutations, numerous SARS-CoV-2 variants have been found, offering a higher risk of infection [20]. A variant of concern is an evolved variant that has a mutation in a specific residue, usually in the spike protein, that may improve the binding affinity of the ACE2 receptor, probably resulting in immune escape or may lead to rapid transmission in human populations [21].
This research compares the presently classified omicron variant to the delta variant in Iraq in order to gain insight into its features and binding affinity with the ACE2 protein. There is evidence that the viral determinant Y505 and the T470–T478 loop in the RBD are necessary for ACE2 to recognize the RBD [22]. T478 is a shared mutation in the delta and omicron variants (Fig. 3). A subunit vaccine for SARS-CoV-2 might be produced using RBD because of its propensity to generate strong neutralizing antibody responses. Antibody mediated immunity may be less effective against the omicron variant because of several alterations in the RBD of spike protein as compared to the delta variant [23] (Fig. 4C, D).
Homology modeling of the SARS-CoV-2 spike protein tetramer identified in Iraq. The homotrimer protein chains A, B, are shown in cartoons in yellow, cyan, and purple. The red color represents the locations of mutations shown in the surface representation. The blue color indicated the insertion mutation. The gray labels indicate shared mutation by the omicron and delta variants. A Homology modeling of the omicron spike protein hCoV-19/Iraq/Sulaimani-1/2022. B Homology modeling of the delta spike protein hCoV-19/Iraq/Sulaimani-11/2021. (Color figure online)
Molecular docking of SARS-CoV-2 RBD-hACE2. The blue color represents human ACE2, and the green color represents RBD monomer. The red color represents the mutation in RBD. The gray labels indicate shared mutation by the omicron and delta variants. A The surface structure of the omicron RBD-ACE2 complex. B The surface structure of the delta RBD-ACE2 complex. C Cartoon of the omicron RBD-ACE2 complex, with the RBD subjected to 15 mutations. D The delta RBD-ACE2 complex represents the RBD with only two mutations. E The interacting amino acid residues are shown as lines in the docking of omicron RBD-ACE2, and the hydrogen bonding interactions are represented by purple dashed lines. F The interacting amino acid residues are shown as lines in the docking of delta RBD-ACE2, and the hydrogen bonding interactions are represented by purple dashed lines. (Color figure online)
The physical properties spike protein
The point isoelectric (pI) of a protein is the pH value at which its net protein charge is zero. A pI value higher than 7 implies an alkaline protein, while a value lower than 7 shows an acidic protein. Despite the fact that the circulated omicron variant has one amino acid fewer than the delta variant, it has a greater molecular weight. In the present study, the omicron variant is expected to have a more alkaline pI than the delta variant. As previously stated [20], the spike protein of both variants showed a high stability score, as a stability score of less than 40 indicates a stable protein structure, while a score of 40 or above indicates an unstable protein structure. The aliphatic index quantifies the volume of a protein occupied by aliphatic amino acids such as alanine on the side chain. The omicron-spike has a higher aliphatic index, so it is more thermostable than the delta variant. The more thermostable a protein is, the higher its aliphatic index [24]. Hydropathicity is a measure of how hydrophobic or hydrophilic amino acids are in a protein sequence. Hydrophilic proteins have a low grand average hydropathicity (GRAVY) score, which indicates that they are polar and water-attracted [24]. A preliminary structural investigation revealed that all SARS-CoV-2 variants share a set of structural characteristics. There were higher percentage of lysine (K), glutamic acid (E) and aspartic acid (D) in the identified OMICRON variant compared to the delta variant. The higher percentage of these amino acids in omicron may contribute to higher salt bridge production (Table S2).
If we compare the omicron spike protein to the detected delta variant, we can see that the omicron spike protein has more hydrophobic amino acids like phenylalanine (F) and isoleucine (I), which may be a consequence of its placement inside the core of the protein. Compared to the circulated delta variant in Iraq, the omicron variant has a reduced proportion of polar amino acids [25]. Because these residues are inside the protein core, the solvent can’t get to them.
Prediction of changes in secondary structural elements
In comparison to delta, omicron variant RBM has a higher percentage of alphahelix structures but fewer extended strands and random coil structures (Table S3) [26]. The expected increase in alpha helices indicates that alpha helices have more mutation resistant than beta strands, which is consistent with prior findings [27]. There is, however, a slight increase in the random coil structure in the RBD of the omicron variant [28].
Prediction of an intrinsically unstructured protein
Viral pathogenicity and infectivity are associated with disordered regions of viral proteins. Residues with predicted disorder values higher than 0.5 are considered intrinsically disordered, whereas residues with predicted disorder scores in the range of 0.2–0.5 are regarded as flexible. Accordingly, the omicron Spike is predicted in this study to contain a lower number of disordered segments than the delta variant according to the PONDR-FIT tool. Similar observations were found in a previous publication [29].
According to an earlier study using the cryo EM structure of the reference virus sequence of Wuhan Hu1, the amino acid residues of Q498-Y505 and T470-F490 loop of spike protein are critical contacting residues that bind the RBD of the virus with ACE2 of the host [22]. In this study, the delta variant RBD exhibited disordered residues in the amino acid residues E471, I472, and Y473 of the RBD. On the other hand, the comparable RBD region of omicron did not contain any disorder residues. This means that there is a possibility of disordered–ordered conversion in spike protein amino acid residues 471–473, which may be significant in the stability effect of spike protein and RBD interaction with ACE2 (Fig. 4E, F).
Protein stability alterations are predicted as a result of mutation
With the exception of the N501Y mutation, which is predicted to increase the stability of the spike protein (Table S4), all amino acid substitutions give the omicron variant the characteristic of being less stable than the delta variant. These observations are consistent with the findings of the previous study [30]. In the RBD of the delta variant, despite the fact that the amino acid substitution mutations of L452R and T478 are tolerable, these mutations affect protein stability and increase the likelihood of developing disease. It is thought that the N501Y mutation of omicron variants, even though it is well tolerated and makes the protein more stable, is a risk mutation for COVID 19 [30].
RBD-ACE2 protein dockin
The receptor recognition mechanism of the SARS-CoV2 virus is crucial for understanding the virus infectivity, pathogenesis, and host range. Due to small mutations in the amino acid composition of RBD, there is a difference in affinity of SARS-CoV-2 variants for ACE2, which leads to differences in ACE2 interactions. Due to its speed and cost-effectiveness, protein–protein docking has become a prominent technique for predicting protein–protein complexes. The RBD of SARS-CoV-2 for both the omicron and delta variants was employed in this investigation for protein–protein docking with human ACE2 (Fig. 4A, B). The docking of the omicron variant revealed that it requires less energy to engage with ACE2 than the delta variant. This means that the omicron variant is more vulnerable to hACE2 attachment than the delta variant, which means there is a greater risk of transmission (Fig. 4).
Almost all of the antibodies that have been identified as being capable of neutralizing SARS-CoV-2 bind to RBD epitopes. A large number of additional antibodies are also in competition for bids with ACE2. Before the emergence of the omicron variant, previous studies showed that a large number of antibodies were still effectively neutralizing SARS-CoV-2 variants [31]. However, recent findings indicate that 85% of previously described neutralizing antibodies are no longer effective against the novel omicron Variant [32]. As shown in the omicron variant of Iraq, the binding residue of RBD has the most mutations, while the delta variant that is found in the area only has two mutations in RBD (Fig. 4E, F).
Conclusion
In this study, the structure, genetics, and dynamic changes that affect total protein stability were investigated in both the omicron and delta variants that were identified in Iraq using a variety of computational tools and molecular methods, with the goal of determining how these changes affect total protein stability. The findings of this study indicated that the circulated omicron variant In Iraq has a large modification in the RBD region, which may contribute to its high binding affinity for the human ACE2 protein. As a result, when compared to the delta variant, this variant may be more contagious in Iraq.
References
Kumar A et al (2021) SARS-CoV-2-specific virulence factors in COVID-19. J Med Virol 93(3):1343–1350
Dawood AA (2022) Increasing the frequency of omicron variant mutations boosts the immune response and may reduce the virus virulence. Microb Pathog 164:105400
Li J et al (2021) The emergence, genomic diversity and global spread of SARS-CoV-2. Nature 600(7889):1–11
Mathieu E et al (2021) A global database of COVID-19 vaccinations. Nat Hum Behav. https://doi.org/10.1038/s41562-021-01122-8
Krause PR et al (2021) Considerations in boosting COVID-19 vaccine immune responses. Lancet 398(10308):1377–1380
Control, C.f.D. and Prevention (2021) Science brief omicron (B. 1.1. 529) variant
Chen J et al (2022) Omicron variant (B.1.1.529): infectivity, vaccine breakthrough, and antibody resistance. J Chem Inf Model 62(2):412–422
Sharma D, Sharma J, Singh A (2021) Exploring the mystery of angiotensin-converting enzyme II (ACE2) in the battle against SARS-CoV-2. J Renin Angioten Aldos Syst. https://doi.org/10.1155/2021/9939929
Abdullah HM et al (2020) Severe refractory COVID-19 patients responding to convalescent plasma; a case series. Ann Med Sur 56:125–127
Sabir DK (2022) Analysis of SARS-COV2 spike protein variants among Iraqi isolates. Gene Rep 26:101420
Gasteiger E et al (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31(13):3784–3788
Aksamentov I et al (2021) Nextclade: clade assignment, mutation calling and quality control for viral genomes. J Open Sour Softw 6(67):3773
Waterhouse A et al (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46(W1):W296–W303
Gurung AB (2020) In silico structure modelling of SARS-CoV-2 Nsp13 helicase and Nsp14 and repurposing of FDA approved antiviral drugs as dual inhibitors. Gene Rep 21:100860
Uversky VN, Gillespie JR, Fink AL (2000) Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 41(3):415–427
Xue B et al (2010) PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta (BBA) 1804(4):996–1010
Capriotti E, Fariselli P, Casadio R (2005) I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res 33:W306–W310
Laskowski RA et al (2020) VarSite: disease variants and protein structure. Protein Sci 29(1):111–119
Desta IT et al (2020) Performance and its limits in rigid body protein-protein docking. Structure 28(9):1071–1081
Corey L et al (2021) SARS-CoV-2 variants in patients with immunosuppression. Mass Med Soc. https://doi.org/10.1056/NEJMsb2104756
Baj A et al (2021) Spike protein evolution in the SARS-CoV-2 delta variant of concern: a case series from Northern Lombardy. Emerg Microbes Infect 10(1):2010–2015
Xu C et al (2021) Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM. Sci Adv 7(1):eabe5575
Hoffmann M et al (2021) The omicron variant is highly resistant against antibody-mediated neutralization–implications for control of the COVID-19 pandemic. Cell 184:2362
Di Rienzo L et al (2021) Characterizing hydropathy of amino acid side chain in a protein environment by investigating the structural changes of water molecules network. Front Mol Biosci 8:626837
Di Rienzo L et al (2021) Characterizing hydropathy of amino acid side chain in a protein environment by investigating the structural changes of water molecules network. Front Mol Biosci 8:2
Combet C et al (2000) NPS@: network protein sequence analysis. Trends Biochem Sci 25(3):147–150
Abrusán G, Marsh JA (2016) Alpha helices are more robust to mutations than beta strands. PLoS Comput Biol 12(12):e1005242
Zhang J et al (2021) Structure of SARS-CoV-2 spike protein. Curr Opin Virol 50:173–182
Kumar S et al (2021) Omicron and delta variant of SARS-CoV-2: a comparative computational study of spike protein. J Med Virol. https://doi.org/10.1002/jmv.27526
Tian F et al (2021) N501Y mutation of spike protein in SARS-CoV-2 strengthens its binding to receptor ACE2. Elife 10:e69091
Du S et al (2021) Structures of SARS-CoV-2 B.1.351 neutralizing antibodies provide insights into cocktail design against concerning variants. Cell Res 31(10):1130–1133
Cao Y et al (2021) Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 602:1–9
Acknowledgements
The authors would like to thank “Shahid Tahir Ali Wali Bag” Hospital and laboratory to provide the samples.
Author information
Authors and Affiliations
Contributions
All the authors equally participated in the study.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethical approval
Verbal approval was taken from the patients to use their disposable oropharyngeal samples. The study was conducted following the research ethical guidelines at the health directorate approved the study (Approval Number 122/10).
Informed consent
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rashid, P.M.A., Salih, G.F. Molecular and computational analysis of spike protein of newly emerged omicron variant in comparison to the delta variant of SARS-CoV-2 in Iraq. Mol Biol Rep 49, 7437–7445 (2022). https://doi.org/10.1007/s11033-022-07545-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07545-4