Abstract
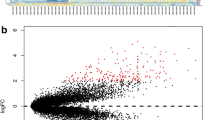
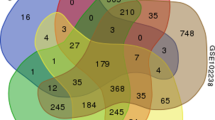
Pancreatic cancer is a uniformly lethal disease that can be difficult to diagnose at its early stage. Thus, our present study aimed to explore the underlying mechanism and identify new targets for this disease. The data GSE16515, including 36 tumor and 16 normal samples were available from Gene Expression Omnibus. Differentially expressed genes (DEGs) were screened out using Robust Multichip Averaging and LIMMA package. Moreover, gene ontology and pathway enrichment analyses were performed to DEGs. Followed with protein–protein interaction (PPI) network construction by STRING and Cytoscape, module analysis was conducted using ClusterONE. Finally, based on PubMed, text mining about these DEGs was carried out. Total 274 up-regulated and 93 down-regulated genes were identified as the common DEGs and these genes were discovered significantly enriched in cell adhesion and extracellular region terms, as well as ECM-receptor interaction pathway. In addition, five modules were screened out from the up-regulated PPI network with none in down-regulated network. Finally, the up-regulated genes, including MIA, MET and CEACAMS, and down-regulated genes, such as FGF, INS and LAPP, had the most references in text mining analysis. Our findings demonstrate that the up- and down-regulated genes play important roles in pancreatic cancer development and might be new targets for the therapy.




Similar content being viewed by others
References
Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C (2007) Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell 1(3):313–323
Li D, Xie K, Wolff R, Abbruzzese JL (2004) Pancreatic cancer. Lancet 363(9414):1049–1057
Buchsbaum DJ, Croce CM (2014) Will detection of microRNA biomarkers in blood improve the diagnosis and survival of patients with pancreatic cancer? JAMA 311(4):363–365
Hidalgo M (2010) Pancreatic cancer. New Engl J Med 362(17):1605–1617
Bardeesy N, DePinho RA (2002) Pancreatic cancer biology and genetics. Nat Rev Cancer 2(12):897–909
Sarkar FH, Banerjee S, Li Y (2007) Pancreatic cancer: pathogenesis, prevention and treatment. Toxicol Appl Pharmacol 224(3):326–336
Asano T, Yao Y, Zhu J, Li D, Abbruzzese JL, Reddy SA (2004) The PI 3-kinase/Akt signaling pathway is activated due to aberrant Pten expression and targets transcription factors NF-κB and c-Myc in pancreatic cancer cells. Oncogene 23(53):8571–8580
Ji Z, Mei FC, Xie J, Cheng X (2007) Oncogenic KRAS activates hedgehog signaling pathway in pancreatic cancer cells. J Biol Chem 282(19):14048–14055
Li M, Zhang Y, Liu Z, Bharadwaj U, Wang H, Wang X, Zhang S, Liuzzi JP, Chang S-M, Cousins RJ (2007) Aberrant expression of zinc transporter ZIP4 (SLC39A4) significantly contributes to human pancreatic cancer pathogenesis and progression. Proc Natl Acad Sci 104(47):18636–18641
Maitra A, Kern SE, Hruban RH (2006) Molecular pathogenesis of pancreatic cancer. Best Pract Res Clin Gastroenterol 20(2):211–226
Pei H, Li L, Fridley BL, Jenkins GD, Kalari KR, Lingle W, Petersen G, Lou Z, Wang L (2009) FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell 16(3):259–266
Team RDC (2005) R: A language and environment for statistical computing. ISBN 3-900051-07-0. R Foundation for Statistical Computing. Vienna, Austria, 2013. url: http://www.R-project.org
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4(2):249–264
Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3(1):3
Hulsen T, de Vlieg J, Alkema W (2008) BioVenn–a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9(1):488
Alvord G, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA (2007) The DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 8:R183
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT (2000) Gene ontology: tool for the unification of biology. Nat Genet 25(1):25–29
Aoki-Kinoshita KF, Kanehisa M (2007) Gene annotation and pathway mapping in KEGG. In: Comparative genomics. Springer, p 71–91
Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C (2013) STRING v9. 1: protein–protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41(D1):D808–D815
Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27(3):431–432
Nepusz T, Yu H, Paccanaro A (2012) Detecting overlapping protein complexes in protein–protein interaction networks. Nat Methods 9(5):471–472
Tsai RT-H, Dai H-J, Lai P-T, Huang C-H (2009) PubMed-EX: a web browser extension to enhance PubMed search with text mining features. Bioinformatics 25(22):3031–3032
Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, Morsberger LA, Latimer C, McLaren S, Lin M-L (2010) The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 467(7319):1109–1113
Hong X, Michalski CW, Kong B, Zhang W, Raggi MC, Sauliunaite D, De Oliveira T, Friess H, Kleeff J (2010) ALCAM is associated with chemoresistance and tumor cell adhesion in pancreatic cancer. J Surg Oncol 101(7):564–569
Parsons JT, Horwitz AR, Schwartz MA (2010) Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Biol 11(9):633–643
Xu Z, Zhang Y, Jiang J, Yang Y, Shi R, Hao B, Zhang Z, Huang Z, Kim JW, Zhang G (2010) Epidermal growth factor induces HCCR expression via PI3 K/Akt/mTOR signaling in PANC-1 pancreatic cancer cells. BMC Cancer 10(1):161
Lee H-O, Mullins S, Franco-Barraza J, Valianou M, Cukierman E, Cheng J (2011) FAP-overexpressing fibroblasts produce an extracellular matrix that enhances invasive velocity and directionality of pancreatic cancer cells. BMC Cancer 11(1):245
Shintani Y, Hollingsworth MA, Wheelock MJ, Johnson KR (2006) Collagen I promotes metastasis in pancreatic cancer by activating c-Jun NH2-terminal kinase 1 and up-regulating N-cadherin expression. Cancer Res 66(24):11745–11753
Baril P, Gangeswaran R, Mahon P, Caulee K, Kocher H, Harada T, Zhu M, Kalthoff H, Crnogorac-Jurcevic T, Lemoine N (2006) Periostin promotes invasiveness and resistance of pancreatic cancer cells to hypoxia-induced cell death: role of the β4 integrin and the PI3k pathway. Oncogene 26(14):2082–2094
Andrianifahanana M, Moniaux N, Schmied BM, Ringel J, Friess H, Hollingsworth MA, Büchler MW, Aubert J-P, Batra SK (2001) Mucin (MUC) gene expression in human pancreatic adenocarcinoma and chronic pancreatitis a potential role of MUC4 as a tumor marker of diagnostic significance. Clin Cancer Res 7(12):4033–4040
Kuespert K, Pils S, Hauck CR (2006) CEACAMs: their role in physiology and pathophysiology. Curr Opin Cell Biol 18(5):565–571
Stoll R, Renner C, Buettner R, Voelter W, Bosserhoff AK, Holak TA (2003) Backbone dynamics of the human MIA protein studied by 15 N NMR relaxation: implications for extended interactions of SH3 domains. Protein Sci 12(3):510–519
Cui JJ, McTigue M, Nambu M, Tran-Dubé M, Pairish M, Shen H, Jia L, Cheng H, Hoffman J, Le P (2012) Discovery of a novel class of exquisitely selective mesenchymal-epithelial transition factor (c-MET) protein kinase inhibitors and identification of the clinical candidate 2-(4-(1-(quinolin-6-ylmethyl)-1 H-[1–3] triazolo [4, 5-b] pyrazin-6-yl)-1 H-pyrazol-1-yl) ethanol (PF-04217903) for the Treatment of Cancer. J Med Chem 55(18):8091–8109
Beenken A, Mohammadi M (2009) The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov 8(3):235–253
Edghill EL, Flanagan SE, Patch A-M, Boustred C, Parrish A, Shields B, Shepherd MH, Hussain K, Kapoor RR, Malecki M (2008) Insulin mutation screening in 1,044 patients with diabetes mutations in the INS gene are a common cause of neonatal diabetes but a rare cause of diabetes diagnosed in childhood or adulthood. Diabetes 57(4):1034–1042
Acknowledgments
This study was supported by the Administration of Traditional Chinese Medicine of Heilongjiang Province (project number ZHY12-Z170). We wish to express our warm thanks to Fenghe(Shanghai) Information Technology Co., Ltd. Their ideas and help gave a valuable added dimension to our research.
Conflict of interest
The authors have declared that no competing interests exist.
Author information
Authors and Affiliations
Corresponding author
Additional information
The Publisher and Editor retract this article in accordance with the recommendations of the Committee on Publication Ethics (COPE). After a thorough investigation we have strong reason to believe that the peer review process was compromised.
About this article
Cite this article
Zhao, LL., Zhang, T., Zhuang, LW. et al. RETRACTED ARTICLE: Uncovering the pathogenesis and identifying novel targets of pancreatic cancer using bioinformatics approach. Mol Biol Rep 41, 4697–4704 (2014). https://doi.org/10.1007/s11033-014-3340-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3340-1