Abstract
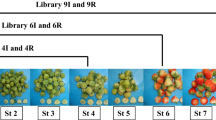
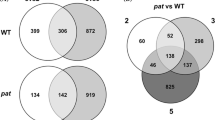
Microarray analysis of genes can provide individual gene-expression profiles and new insights for elucidating biological mechanisms responsible for fruit development. To obtain an overall view on expression profiles of metabolism-related genes involved in fruit development of table and wine grapes, a microarray system comprising 15,403 ESTs was used to compare the expressed genes. The expression patterns from the microarray analysis were validated with quantitative real-time polymerase chain reaction analysis of 18 selected genes of interest. During the entire fruit development stage, 2,493 genes exhibited at least 2.0-fold differences in expression levels with 1,244 genes being up-regulated and 1,249 being down-regulated. Following gene ontology analysis, only 929 differentially expressed (including 403 up-regulated and 526 down-regulated) genes were annotated in table and wine grapes. These differentially expressed genes were found to be mainly involved in carbohydrate metabolism, biosynthesis of secondary metabolites as well as energy, lipid and amino acid metabolism via KEGG. Our results provide new insights into the molecular mechanisms and expression profiles of genes in the fruit development stage of table and wine grapes.





Similar content being viewed by others
Abbreviations
- GO:
-
Gene ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- qRT-PCR:
-
Quantitative reverse transcription-polymerase chain reaction
- MDH:
-
Malate dehydrogenase
- TCA cycle:
-
Tricarboxylic acid cycle
References
Peng FY, Reid KE, Liao N, Schlosser J, Lijavetzky D, Holt R, Martínez Zapater JM, Jones S, Marra M, Bohlmann J, Lund ST (2007) Generation of ESTs in Vitis vinifera wine grape (Cabernet Sauvignon) and table grape (Muscat Hamburg) and discovery of new candidate genes with potential roles in berry development. Gene 402:40–50
The French-Italian Public Consortium for Grapevine Genome Characterization (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–468
Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436:344–356
Tuskan GA, DiFazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313:1595–1604
Boss PK, Vivier M, Matsumoto S, Dry IB, Thomas MR (2001) A cDNA from grapevine (Vitis vinifera L.), which shows homology to AGAMOUS and SHATTERPROOF, is not only expressed in flowers but also throughout berry development. Plant Mol Biol 45:541–553
Manganaris GA, Ziliotto F, Rasori A, Bonghi C, Ramina A, Tonutti P (2010) A comparative transcriptomic approach to elucidate common and divergent mechanisms involved in apricot and peach fruit development and ripening. Acta Hortic 862:577–582
Wang Y, Li J, Yang J, Xia R (2011) Expression of lycopene cyclase genes and their regulation on downstream carotenoids during fruit maturation of Guoqing No. 1 Satsuma mandarin and Cara Cara navel orange. Sci Hortic 127:267–274
Zhang Y, Li P, Cheng L (2010) Developmental changes of carbohydrates, organic acids, amino acids, and phenolic compounds in ‘Honeycrisp’ apple flesh. Food Chem 123:1013–1018
Deluc LG, Grimplet J, Wheatley MD, Tillett RD, Quilici DR, Osborne C, Schooley DA, Schlauch KA, Cushman JC, Cramer GR (2007) Transcriptomic and metabolite analyses of Cabernet Sauvignon grape berry development. BMC Genom 8:492
Singh RK, Sane VU, Misra A, Ali SA, Nath P (2010) Differential expression of the mango alcohol dehydrogenase gene family during ripening. Phytochemistry 71:1485–1494
Duggan DJ, Bittner M, Chen Y, Meltzer P, Trent JM (1999) Expression profiling using cDNA microarrays. Nat Genet 21:10–14
Cuzin M (2001) DNA chips: a new tool for genetic analysis and diagnostics. Transfus Clin Biol 8:291–296
Ruan Y, Gilmore J, Conner T (1998) Towards Arabidopsis genome analysis: monitoring expression profiles of 1400 genes using cDNA microarrays. Plant J 15:821–833
Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K (2001) Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13:61–72
Schena M, Shalon D, Heller R, Chai A, Brown PO, Davis RW (1996) Parallel human genome analysis: microarray based expression monitoring of 1000 genes. Proc Natl Acad Sci USA 93:10614–10619
Waters DLE, Holton TA, Ablett EM, Lee LS, Henry RJ (2005) cDNA microarray analysis of developing grape (Vitis vinifera cv. Shiraz) berry skin. Funct Integr Genomics 5:40–58
Ali MB, Howard S, Chen S, Wang Y, Yu O, Kovacs LG, Qiu W (2011) Berry skin development in Norton grape: distinct patterns of transcriptional regulation and flavonoid biosynthesis. BMC Plant Biol 11:7
Lund ST, Peng FY, Nayar T, Reid KE, Schlosser J (2008) Gene expression analyses in individual grape (Vitis vinifera L.) berries during ripening initiation reveal that pigmentation intensity is a valid indicator of developmental staging within the cluster. Plant Mol Biol 68:301–315
Abdullahi I, Rott M (2009) Microarray immunoassay for the detection of grapevine and tree fruit viruses. J Virol Methods 160:90–100
Camps C, Kappel C, Lecomte P, Leon C, Gomes E, Coutos-Thevenot P, Delrot S (2010) A transcriptomic study of grapevine (Vitis vinifera cv. Cabernet-Sauvignon) interaction with the vascular ascomycete fungus Eutypa lata. J Exp Bot 61:1719–1737
Legay G, Marouf E, Berger D, Neuhaus JM, Mauch-Mani B, Slaughter A (2011) Identification of genes expressed during the compatible interaction of grapevine with Plasmopara viticola through suppression subtractive hybridization (SSH). Eur J Plant Pathol 129:281–301
Cramer GR, Ergül A, Grimplet J, Tillett RL, Tattersall EAR, Bohlman MC, Vincent D, Sonderegger J, Evans J, Osborne C, Quilici D, Schlauch KA, Schooley DA, Cushman JC (2007) Water and salinity stress in grapevines: early and late changes in transcript and metabolite profiles. Funct Integr Genomics 7:111–134
Daldoul S, Guillaumie S, Reustle GM, Krczal G, Ghorbel A, Delrot S, Mliki A, Hofer MU (2010) Isolation and expression analysis of salt induced genes from contrasting grapevine (Vitis vinifera L.) cultivars. Plant Sci 179:489–498
Tillett RL, Ergül A, Albion RL, Schlauch KA, Cramer GR, Cushman JC (2011) Identification of tissue-specific, abiotic stress-responsive gene expression patterns in wine grape (Vitis vinifera L.) based on curation and mining of large-scale EST data sets. BMC Plant Biol 11:86
Figueiredo A, Monteiro F, Fortes AM, Bonow-Rex M, Zyprian E, Sousa L, Pais MS (2012) Cultivar-specific kinetics of gene induction during downy mildew early infection in grapevine. Funct Integr Genomics 12(2):379–386
Reid KR, Olsson N, Schlosser J, Peng F, Lund ST (2006) An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol 6:27
Saavedra L, Barbas C (2003) Validated capillary electrophoresis method for small-anions measurement in wines. Electrophoresis 24:2235–2243
Alba R, Fei Z, Payton P, Liu Y, Moore SL, Debbie P, Cohn J, D’Ascenzo M, Gordon JS, Rose JKC, Martin G, Tanksley SD, Bouzayen M, Jahn MM, Giovannoni J (2004) ESTs cDNA microarrays and gene expression profiling: tools for dissecting plant physiology and development. Plant J 39:697–714
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95:14863–14868
Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Kanehisa M, Goto S (2000) KEGG: Kyoto encyclopaedia of genes and genomes. Nucleic Acids Res 28:27–30
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:D480–D484
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using realtime quantitative PCR and the 2△△C(T) method. Methods 25:402–408
Clarke JD, Zhu T (2006) Microarray analysis of the transcriptome as a stepping stone towards understanding biological systems: practical considerations and perspectives. Plant J 45:630–650
Soglio V, Costa F, Molthoff JW, Weemen-Hendriks WMJ, Schouten HJ, Gianfranceschi L (2009) Transcription analysis of apple fruit development using cDNA microarrays. Tree Genet Genomes 5:685–698
Naoumkina MA, Modolo LV, Huhman DV (2010) Genomic and coexpression analyses predict multiple genes involved in triterpene saponin biosynthesis in Medicago truncatula. Plant Cell 22:850–866
Cui GH, Huang LQ, Tang XJ (2011) Candidate genes involved in tanshinone biosynthesis in hairy roots of Salvia miltiorrhiza revealed by cDNA microarray. Mol Biol Rep 38:2471–2478
Manrique-Trujillo SM, Ramírez-López AC, Ibarra-Laclette E, Gómez-Lim MA (2007) Identification of genes differentially expressed during ripening of banana. J Plant Physiol 164(8):1037–1050
Lee DH, Kang SG, Suh SG, Byun JK (2003) Purification and characterization of a β-galactosidase from peach (Prunus persica). Mol Cells 15:68–74
Sekine D, Munemura I, Gao M, Mitsuhashi W, Toyomasu T, Murayama H (2006) Cloning of cDNAs encoding cell-wall hydrolases from pear (Pyrus communis) fruit and their involvement in fruit softening and development of melting texture. Physiol Plant 126:163–174
Murayama H, Arikawa M, Sasaki YD, Cin V, Mitsuhashi W, Toyomasu T (2009) Effect of ethylene treatment on expression of polyuronide-modifying genes and solubilization of polyuronides during ripening in two peach cultivars having different softening characteristics. Postharvest Biol Technol 52(2):196–201
Sulova Z, Baran R, Farkas V (2001) Release of complexed xyloglucan endotransglycosylase (XET) from plant cell walls by a transglycosylation reaction with xyloglucan-derived oligosaccharides. Plant Physiol Biochem 39(11):927–932
Alaycdn-Luaces P, Pagano EA, Mroginski LA, Sozzi GO (2010) Activity levels of six glycoside hydrolases in apple fruit callus cultures depend on the type and concentration of carbohydrates supplied and the presence of plant growth regulators. Plant Cell Tissue Organ Cult 101(1):1–10
Ortega-García F, Peragón J (2009) Phenylalanine ammonia-lyase, polyphenol oxidase, and phenol concentration in fruits of Olea europaea L. cv. Picual, Verdial, Arbequina, and Frantoio during ripening. J Agric Food Chem 57(21):10331–10340
Achnine L, Blancaflor EB, Rasmussen S, Dixon RA (2004) Colocalization of l-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis. Plant Cell 16:3098–3109
Salentijn EMJ, Aharoni A, Schaart JG, Boone MJ, Krens FA (2003) Differential gene expression analysis of strawberry cultivars that differ in fruit-firmness. Physiol Plant 118:571–578
Wang Y, Li J, Xia RX (2010) Expression of chalcone synthase and chalcone isomerase genes and accumulation of corresponding flavonoids during fruit maturation of Guoqing No. 4 satsuma mandarin (Citrus unshiu Marcow). Sci Hortic 125(2):110–116
Zheng YJ, Tian L, Liu HT, Pan QH, Zhan JC, Huang WD (2009) Sugars induce anthocyanin accumulation and flavanone 3-hydroxylase expression in grape berries. Plant Growth Regul 58(3):251–260
Fujita A, Goto-Yamamoto N, Aramaki I, Hashizume K (2006) Organ-specific transcription of putative flavonol synthase genes of grapevine and effects of plant hormones and shading on flavonol biosynthesis in grape berry skins. Biosci Biotechnol Biochem 70(3):632–638
Wang Z, Zhao FX, Zhao X, Ge H, Chai LJ, Chen SW, Perl A, Ma HQ (2012) Proteomic analysis of berry-sizing effect of GA3 on seedless Vitis vinifera L. Proteomics 12:86–94
Moore S, Vrebalov J, Payton P, Giovannoni J (2002) Use of genomics tools to isolate key ripening genes and analyse fruit maturation in tomato. J Exp Bot 53:2023–2030
Robles P, Pelaz S (2005) Flower and fruit development in Arabidopsis thaliana. Int J Dev Biol 49:633–643
Xu BY, Su W, Liu JH, Wang JB, Jin ZQ (2007) Differentially expressed cDNAs at the early stage of banana ripening identified by suppression subtractive hybridization and cDNA microarray. Planta 226(2):529–539
Yao Y, Li M, Liu Z, Hao Y, Zhai H (2007) A novel gene screened by cDNA-AFLP approach contributes to lowering the acidity of fruit in apple. Plant Physiol 45:139–145
Miller SS, Driscoll BT, Gregerson RG (1998) Alfalfa malate dehydrogenase (MDH): molecular cloning and characterization of five different forms reveals a unique nodule enhance MDH. Plant J 15:173–184
Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor MdMYB10. Plant J 49:414–427
Hedden P, Phillips AL (2000) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5:523–530
Lee Y, Yu G, Seo YS, Han SE, Choi Y, Kim D, Mok I, Kim WT, Sung S (2007) Microarray analysis of apple gene expression engaged in early fruit development. Plant Cell Rep 26:917–926
Acknowledgments
This project was supported by the Fundamental Research Funds for the Central Universities (No. KYJ200909) and the Program of NCET (No. NCET08-0796).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2014_3311_MOESM1_ESM.doc
Additional File 1. Percentage distribution of up/down-regulated differentially expressed genes involved in biological process, molecular functions and cellular component annotation. (DOC 441 kb)
11033_2014_3311_MOESM2_ESM.cdt
Additional File 2. 2,493 differentially expressed genes with two fold increase or decrease detected by microarray between fruits of table and wine grape. This file can be recognized via Tree View 1.6 software. The clustering diagram shows that the up- or down regulated genes between table and wine grape were divided into two groups, with genes having similar expression profile were clustered into the sub-groups. (CDT 299 kb)
11033_2014_3311_MOESM3_ESM.pdf
Additional File 3. The metabolic pathways map corresponding to up/down-regulated differentially expressed genes encoding enzymes. The red lines shown in metabolic pathway figure represent the up-regulated differentially expressed genes, which were with high expression in table grape, the green ones represent the down-regulated genes, which were with high expression in wine grape. (PDF 189 kb)
Rights and permissions
About this article
Cite this article
Wang, X., Kayesh, E., Han, J. et al. Microarray analysis of differentially expressed genes engaged in fruit development between table and wine grape. Mol Biol Rep 41, 4397–4412 (2014). https://doi.org/10.1007/s11033-014-3311-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3311-6