Abstract
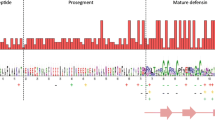
Defensins are endogenous peptides with cysteine-rich antimicrobial ability that contribute to host defence against bacterial, fungal and viral infections. There are three subfamilies of defensins in primates: α, β and θ-defensins. α-defensins are most present in neutrophils and Paneth cells; β-defensins are involved in protecting the skin and the mucous membranes of the respiratory, genitourinary and gastrointestinal tracts; and θ-defensins are physically distinguished as the only known fully-cyclic peptides of animal origin, which are first isolated from rhesus macaques. All three kinds of defensins have six conserved cysteines, three intramolecular disulfide bonds, a net positive charge, and β-sheet regions. α and θ-defensins are closely related, comparative amino acid sequences showed that the difference between them is that θ-defensins have an additional stop codon limits the initial defensin domain peptides to 12 residues. Humans, chimpanzees and gorillas do not produce θ-defensin peptides due to a premature stop codon present in the signal sequence of all θ-defensin pseudogenes. By using comprehensive computational searches, here we report the discovery of complete repertoires of the α and θ-defensin gene family in ten primate species. Consistent with previous studies, our phylogenetic analyses showed all primate θ-defensins evident formed one distinct clusters evolved from α-defensins. β-defensins are ancestors of both α and θ-defensins. Human has two copies of DEFA1 and DEFT1P, and two extra DEFA3 and DEFA10P genes compared with gorilla. As different primates inhabit in quite different ecological niches, the production of species-specific α and θ-defensins and these highly evolved θ-defensins in old world monkeys would presumably allow them to better respond to the specific microbial challenges that they face.



Similar content being viewed by others
References
Zou J, Mercier C, Koussounadis A, Secombes C (2007) Discovery of multiple beta-defensin like homologues in teleost fish. Mol Immunol 44(4):638–647
Hellgren O, Ekblom R (2010) Evolution of a cluster of innate immune genes (beta-defensins) along the ancestral lines of chicken and zebra finch. Immunome Res 6:3. doi:10.1186/1745-7580-6-3
Hughes A (1999) Evolutionary diversification of the mammalian defensins. Cell Mol Life Sci 56(1–2):94–103
Aldred PM, Hollox EJ, Armour JA (2005) Copy number polymorphism and expression level variation of the human α-defensin genes DEFA1 and DEFA3. Hum Mol Genet 14(14):2045–2052
Ganz T (1999) Defensins and host defense. Science 286(5439):420–421
Tang Y-Q, Yuan J, Ösapay G, Ösapay K, Tran D, Miller CJ, Ouellette AJ, Selsted ME (1999) A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated α-defensins. Science 286(5439):498–502
Ganz T (2003) Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol 3(9):710–720
Lehrer RI, Ganz T (2002) Defensins of vertebrate animals. Curr Opin Immunol 14(1):96–102
Conibear AC, Rosengren KJ, Daly NL, Henriques ST, Craik DJ (2013) The cyclic cystine ladder in theta-defensins is important for structure and stability, but not antibacterial activity. J Biol Chem 288(15):10830–10840. doi:10.1074/jbc.M113.451047
Nguyen TX, Cole AM, Lehrer RI (2003) Evolution of primate θ-defensins: a serpentine path to a sweet tooth. Peptides 24(11):1647–1654
Liu L, Zhao C, Heng HH, Ganz T (1997) The human beta-defensin-1 and alpha-defensins are encoded by adjacent genes: two peptide families with differing disulfide topology share a common ancestry. Genomics 43(3):316–320. doi:10.1006/geno.1997.4801
Patil A, Hughes AL, Zhang G (2004) Rapid evolution and diversification of mammalian alpha-defensins as revealed by comparative analysis of rodent and primate genes. Physiol Genomics 20(1):1–11. doi:10.1152/physiolgenomics.00150.2004
Chen H, Xu Z, Peng L, Fang X, Yin X, Xu N, Cen P (2006) Recent advances in the research and development of human defensins. Peptides 27(4):931–940
Klotman ME, Chang TL (2006) Defensins in innate antiviral immunity. Nat Rev Immunol 6(6):447–456
Daher K, Selsted M, Lehrer R (1986) Direct inactivation of viruses by human granulocyte defensins. J Virol 60(3):1068–1074
Ghosh D, Porter E, Shen B, Lee SK, Wilk D, Drazba J, Yadav SP, Crabb JW, Ganz T, Bevins CL (2002) Paneth cell trypsin is the processing enzyme for human defensin-5. Nat Immunol 3(6):583–590
Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL (2003) Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature 422(6931):522–526
Mackewicz CE, Yuan J, Tran P, Diaz L, Mack E, Selsted ME, Levy JA (2003) [alpha]-Defensins can have anti-HIV activity but are not CD8 cell anti-HIV factors. AIDS 17(14):F23–F32
Chang TL, Vargas J, DelPortillo A, Klotman ME (2005) Dual role of α-defensin-1 in anti–HIV-1 innate immunity. J Clin Investig 115(3):765–773
Yount NY, Wang M, Yuan J, Banaiee N, Ouellette AJ, Selsted ME (1995) Rat neutrophil defensins. Precursor structures and expression during neutrophilic myelopoiesis. J Immunol 155(9):4476–4484
Ferguson L, Browning B, Huebner C, Petermann I, Shelling A, Demmers P, McCulloch A, Gearry R, Barclay M, Philpott M (2008) Single nucleotide polymorphisms in human Paneth cell defensin A5 may confer susceptibility to inflammatory bowel disease in a New Zealand Caucasian population. Dig Liver Dis 40(9):723–730
Aldred PM, Hollox EJ, Armour JA (2005) Copy number polymorphism and expression level variation of the human alpha-defensin genes DEFA1 and DEFA3. Hum Mol Genet 14(14):2045–2052. doi:10.1093/hmg/ddi209
Cole AM, Hong T, Boo LM, Nguyen T, Zhao C, Bristol G, Zack JA, Waring AJ, Yang OO, Lehrer RI (2002) Retrocyclin: a primate peptide that protects cells from infection by T- and M-tropic strains of HIV-1. Proc Natl Acad Sci USA 99(4):1813–1818. doi:10.1073/pnas.052706399
Wang W, Cole AM, Hong T, Waring AJ, Lehrer RI (2003) Retrocyclin, an antiretroviral theta-defensin, is a lectin. J Immunol 170(9):4708–4716
Tran D, Tran PA, Tang YQ, Yuan J, Cole T, Selsted ME (2002) Homodimeric theta-defensins from rhesus macaque leukocytes: isolation, synthesis, antimicrobial activities, and bacterial binding properties of the cyclic peptides. J Biol Chem 277(5):3079–3084. doi:10.1074/jbc.M109117200
Leonova L, Kokryakov VN, Aleshina G, Hong T, Nguyen T, Zhao C, Waring AJ, Lehrer RI (2001) Circular minidefensins and posttranslational generation of molecular diversity. J Leukoc Biol 70(3):461–464
Lehrer RI, Cole AM, Selsted ME (2012) θ-defensins: cyclic peptides with endless potential. J Biol Chem 287(32):27014–27019
Tongaonkar P, Tran P, Roberts K, Schaal J, Ösapay G, Tran D, Ouellette AJ, Selsted ME (2011) Rhesus macaque θ-defensin isoforms: expression, antimicrobial activities, and demonstration of a prominent role in neutrophil granule microbicidal activities. J Leukoc Biol 89(2):283–290
Garcia AE, Osapay G, Tran PA, Yuan J, Selsted ME (2008) Isolation, synthesis, and antimicrobial activities of naturally occurring theta-defensin isoforms from baboon leukocytes. Infect Immun 76(12):5883–5891. doi:10.1128/IAI.01100-08
Ouellette A, Selsted M (1996) Paneth cell defensins: endogenous peptide components of intestinal host defense. FASEB J 10(11):1280–1289
Yeh R-F, Lim LP, Burge CB (2001) Computational inference of homologous gene structures in the human genome. Genome Res 11(5):803–816
Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268(1):78–94
Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12(4):656–664
Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31(13):3497–3500
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Zuckerkandl E, Pauling L (1965) Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ (eds) Evolving genes and proteins. Academic Press, New York, pp 97–166
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39(4):783–791
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17(6):368–376
Hughes AL, Yeager M (1997) Coordinated amino acid changes in the evolution of mammalian defensins. J Mol Evol 44(6):675–682
Liu L, Zhao C, Heng HH, Ganz T (1997) The human β-defensin-1 and α-defensins are encoded by adjacent genes: two peptide families with differing disulfide topology share a common ancestry. Genomics 43(3):316–320
Hughes AL, Piontkivska H (2008) Functional diversification of the toll-like receptor gene family. Immunogenetics 60(5):249–256. doi:10.1007/s00251-008-0283-5
Piertney SB, Oliver MK (2006) The evolutionary ecology of the major histocompatibility complex. Heredity (Edinb) 96(1):7–21. doi:10.1038/sj.hdy.6800724
Hoffmann JA, Kafatos FC, Janeway CA, Ezekowitz R (1999) Phylogenetic perspectives in innate immunity. Science 284(5418):1313–1318
Hughes AL (2002) Natural selection and the diversification of vertebrate immune effectors. Immunol Rev 190(1):161–168
Acknowledgments
This work was supported by China Agriculture Research System (CARS-41), and the Program from Sichuan Province (2011NZ0099-7 and 2011NZ0073).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Diyan Li, Long Zhang and Huadong Yin have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, D., Zhang, L., Yin, H. et al. Evolution of primate α and θ defensins revealed by analysis of genomes. Mol Biol Rep 41, 3859–3866 (2014). https://doi.org/10.1007/s11033-014-3253-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3253-z