Abstract
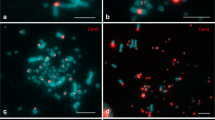
It is well known that transcriptionally inactive rRNA genes are correlated with DNA hyper-methylation and histone hypo-methylation and there is clear evidence in humans that DNA and histone modification which alter chromatin structure are related to chromosome fragility. Very little is known about the biological cause of 45S rDNA fragility. In this report we characterized the chromosome breakage or gap associated with 45S rDNA in a fish species Channa punctatus. The rDNA mapping in C. punctatus, showed many chromosome breakages or gap formations, and all occurred exclusively in the 45S rDNA sites in anterior kidney cells. We observed that the number of chromosomes plus chromosome fragments was often more than the expected 32 in most cells. Total 67 % metaphase spread showed the expected or normal 32 chromosomes, while 33 % metaphase spread showed 33 and/or 34 chromosomes and/or chromosome fragments. The chromosome lesions observed in this study are very similar cytologically to that of fragile sites observed in human chromosomes. Possible causes for the spontaneous expression of fragile sites and their potential biological significance are also discussed in present report.



Similar content being viewed by others
References
Robert-Fortel I, Junera HR, Geraud G, Hernadez-Verdun D (1993) Three-dimensional organization of the ribosomal genes and Ag-NOR proteins during interphase and mitosis in PtK 1 cells studied by confocal microscopy. Chromosoma 102:146–157
Earley K, Lawrence RJ, Pontes O, Reuther R, Enciso AJ et al (2006) Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Gene Dev 20:1283–1293
Wang Y (2006) Chromatin structure of human chromosomal fragile sites. Cancer Lett 232:70–78
Rhichards RI (2001) Fragile and unstable chromosomes in cancer: causes and consequences. Trends Genet 17:339–345
Glover TW (2006) Common fragile sites. Cancer Lett 232:4–12
Magenis RE, Hecht F, Lovrien EW (1970) Heritable fragile site on chromosome 16: probable localization of haptoglobin locus in man. Science 170:85–87
Buttel I, Fechter A, Schwab M (2004) Common fragile sites and cancer: targeted cloning by insertional mutagenesis. Ann NY Acad Sci 1028:14–27
Sutherland GR, Baker E, Richards RI (1998) Fragile sites still breaking. Trends Genet 14:501–506
Huebner K, Croce CM (2001) FRA3B and other common fragile sites: the weakest links. Nat Rev Cancer 1:214–221
Popescu NC (2003) Genetic alterations in cancer as a result of breakage at fragile sites. Cancer Lett 192:1–17
Handt O, Baker E, Dayan S, Gartler SM, Woollatt E et al (2008) Analysis of replication timing at the FRA10B and FRA16B fragile site loci. Chromosome Res 8:677–688
Ng PKL, Lim Kelvin KP (1990) Snakeheads (Pisces: Channidae): Natural History, Biology and economic Importance. In Chou LM, Ng PKL (eds.) Essays in Zoology, National University of Singapore, pp 127-152. I
Diniz D, Bertollo LAC (2003) Karyotypic studies on Hoplerythrinus unitaeniatus (Pisces, Erythrinidae) populations: a biodiversity analysis. Caryologia 56:303–313
Rishi KK, Haobam MS (1990) A chromosomal study on four species of snake heads (Ophiocephalidae: pisces) with comments on their karyotypic evolution. Caryologia 43:163–167
Bertollo LAC, Takahashi CS, Moreira-Filho O (1978) Cytotaxonomic consideration on Hoplias lacrdae (Pisces, Erythrinidae). Braz J Genet 1:103–120
Singh M, Kumar R, Nagpure NS, Kushwaha B, Mani I, Lakra WS (2010) Nucleotide Sequences and Chromosomal Localisation of 18S and 5SrDNA in N. hexagonolepis using dual colour FISH. Zool Sci 27:709–716
Zardoya R, Meyer A (1996) Evolutionary relationships of the coela-canth, lungfishes, and tetrapods based on the 28S ribosomal RNA gene. Proc Natl Acad Sci, USA 93:5449–5454
Winterfeld G, Roser M (2007) Deposition of ribosomal DNAs in the chromosome of perennial oats (Poaceae: aveneae). Bot J Linnean Soc 155:193–210
Singh M, Kumar R, Nagpure NS, Kushwaha B, Gond I, Lakra WS (2009) Chromosomal Localization of 18S and 5S rDNA using FISH in genus Tor (Pisces, Cyprinidae). Genetica 137:245–252
Santi-Rampazzo AP, Nishiyama PB, Ferreira PEB, Martins-Santos IC (2008) Intrapopulational polymorphism of nucleolus organizer regions in Serrapinnus notomelas (Characidae, Cheirodontinae) from the Parana River. J Fish Biol 72:1236–1243
Siroky J, Zluvova J, Riha K, Shippen DE, Vyskot B (2003) Rearrangements of ribosomal DNA clusters in late generation telomerase-deficient Arabidopsis. Chromosoma 112:116–123
Butler DK (1992) Ribosomal DNA is a site of chromosome breakage in aneuploid strains of Neurospora. Genetics 131:581–592
Mishmar D, Rahat A, Scherer SW, Nyakatura G, Hinzmann B et al (1998) Molecular characterization of a common fragile site (FRA7H) on human chromosome 7 by the cloning of a simian virus 40 integration site. Proc Natl Acad Sci USA 95:8141–8146
Ried K, Finnis M, Hobson L, Mangelsdorf M, Dayan S et al (2000) Common chromosomal fragile site FRA16D sequence: identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum Mol Genet 9:1651–1663
Arlt MF, Xu B, Durkin SG, Casper AM, Kastan MB et al (2004) BRCA1 is required for common-fragile-site stability via its G2/M checkpoint function. Mol Cell Biol 24:6701–6709
Wang YH, Griffith J (1996) Methylation of expanded CCG triplet repeat DNA from fragile X syndrome patients enhances nucleosome exclusion. J Biol Chem 271:22937–22940
Yu S, Mangelsdorf M, Hewett D, Hobson L, Baker E et al (1997) Human chromosomal fragile site FRA16B is an amplified AT-rich minisatellite repeat. Cell 88:367–374
Fan Q, Yao MC, Journals O (2000) A long stringent sequence signal for programmed chromosome breakage in Tetrahymena thermophila. Nucl Acids Res 28:895–900
Yao MC (1991) Ribosomal RNA gene amplification in Tetrahymena may be associated with chromosome breakage and DNA elimination. Cell 24:765–774
Ramanathan B, Smerdon MJ (1989) Enhanced DNA repair synthesis in hyperacetylated nucleosomes. J Biol Chem 264:11026–11034
Gu Y, Shen Y, Gibbs RA, Nelson DL (1996) Identification of FMR2, a novel gene associated with the FRAXE CCG repeat and CpG island. Nat Genet 13:109–113
Pomponi MG, Neri G (1994) Butyrate and acetyl-carnitine inhibit the cytogenetic expression of the fragile X in vitro. Am J Med Genet 51:447–450
Lawrence RJ, Earley K, Pontes O, Silva M, Chen ZJ et al (2004) A concerted DNA methylan/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol Cell 13:599–609
McStay B (2006) Nucleolar dominance: a model for rRNA gene silencing. Gene Dev 20:1207–1214
Huang Y (2002) Transcriptional silencing in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Nucl Acids Res 30:1465–1482
Durand-Dubief M, Sinha I, Fagerstrom-Billai F, Bonilla C, Wright A et al (2007) Specific functions for the fission yeast Sirtuins Hst2 and Hst4 in gene regulation and retrotransposon silencing. EMBO J 26:2477–2488
Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA et al (2008) Response to comment on ‘HST2 mediates SIR2-independent life-span extension by calorie restriction’. Science 312:1312
Sutherland GR, Jacky P, Baker E (1984) Heritable fragile sites on human chromosomes. XI. Factors affecting expression of fragile sites at 10q25, 16q22, and 17p12. Am J Hum Genet 36:110–122
Gericke GS (1999) Chromosomal fragility may be indicative of altered higher order DNA organization as the underlying genetic diathesis in complex neuro-behavioural disorders. Med Hypotheses 52:201–208
Coquelle A, Pipiras E, Toledo F, Buttin G, Debatisse M (1997) Expression of fragile sites triggers intra-chromosomal mammalian gene amplification and sets boundaries to early amplicons. Cell 89:215–225
Rassool FV, McKeithan TW, Neilly ME, Melle EV, Espinosa R III et al (1991) Preferential integration of marker DNA into the chromosomal fragile site at 3p14: an approach to cloning fragile sites. Proc Natl Acad Sci USA 88:6657–6661
Author information
Authors and Affiliations
Corresponding author
Additional information
Mamta Singh and Anindya Sundar Barman contributed equally.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Singh, M., Barman, A.S. Chromosome breakages associated with 45S ribosomal DNA sequences in spotted snakehead fish Channa punctatus . Mol Biol Rep 40, 723–729 (2013). https://doi.org/10.1007/s11033-012-2112-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-2112-z