Abstract
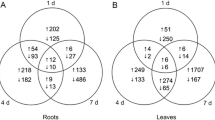
Chickpea (Cicer arietinum L.) is an important pulse crop grown mainly in the arid and semi-arid regions of the world. Due to its taxonomic proximity with the model legume Medicago truncatula and its ability to grow in arid soil, chickpea has its unique advantage to understand how plant responds to drought stress. In this study, an oligonucleotide microarray was used for analyzing the transcriptomic profiles of unigenes in leaf and root of chickpea seedling under drought stress, respectively. Microarray data showed that 4,815 differentially expressed unigenes were either ≥2-fold up- or ≤0.5-fold down-regulated in at least one of the five time points during drought stress. 2,623 and 3,969 unigenes were time-dependent differentially expressed in root and leaf, respectively. 110 pathways in two tissues were found to respond to drought stress. Compared to control, 88 and 52 unigenes were expressed only in drought-stressed root and leaf, respectively, while nine unigenes were expressed in both the tissues. 1,922 function-unknown unigenes were found to be remarkably regulated by drought stress. The expression profiles of these time-dependent differentially expressed unigenes were useful in furthering our knowledge of molecular mechanism of plant in response to drought stress.







Similar content being viewed by others
References
Xiong L, Wang RG, Mao G, Koczan JM (2006) Identification of drought tolerance determinants by genetic analysis of root response to drought stress and abscisic acid. Plant Physiol 142:1065–1074
Bray EA (1997) Plant responses to water deficit. Trends Plant Sci 2:48–54
Ingram J, Bartels D (1996) The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol 47:377–433
Shinozaki K, Yamaguchi-Shinozaki K (1996) Molecular responses to drought and cold stress. Curr Opin Biotechnol 7:161–167
Campalans A, Messeguer R, Goday A, Pages M (1999) Plant responses to drought, from ABA signal transduction events to the action of the induced proteins. Plant Physiol Biochem 37:327–340
Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781–803
Database FAO (2010) http://faostat.fao.org. Accessed Aug and Sep 1997
Jayashree B, Hutokshi KB, Sanjeev S, Jonathan HC (2005) A legume genomics resource: the chickpea root expressed sequence tag database. Electron J Biotechnol 8:128–133
Romo S, Labrador E, Dopico B (2001) Water stress-regulated gene expression in Cicer arietinum seedlings and plants. Plant Physiol Biochem 39:1017–1026
Boominathan P, Shukla R, Kumar A, Manna D, Negi D, Verma PK, Chattopadhyay D (2004) Long term transcript accumulation during the development of dehydration adaptation in Cicer arietinum. Plant Physiol 135:1608–1620
Mantri N, Ford R, Coram T, Pang E (2007) Transcriptional profiling of chickpea genes differentially regulated in response to high-salinity, cold and drought. BMC Genomics 8:303
Molina C, Rotter B, Horres R, Udupa S, Besser B, Bellarmino L, Baum M, Matsumura H, Terauchi R, Kahl G, Winter P (2008) SuperSAGE: the drought stress-responsive transcriptome of chickpea roots. BMC Genomics 9:553
Gao WR, Wang XS, Liu QY, Peng H, Chen C, Li JG, Zhang JS, Hu SN, Ma H (2008) Comparative analysis of ESTs in response to drought stress in chickpea (C. arietinum L.). Biochem Biophys Res Commun 376:578–583
Andersson A, Keskitalo J, Sjodin A, Bhalerao R, Sterky F, Wissel K, Tandre K, Aspeborg H, Moyle R, Ohmiya Y, Bhalerao R, Brunner A, Gustafsson P, Karlsson J, Lundeberg J, Nilsson O, Sandberg G, Strauss S, Sundberg B, Uhlen M, Jansson S, Nilsson P (2004) A transcriptional timetable of autumn senescence. Genome Biol 5:R24
Jain D, Chattopadhyay D (2010) Analysis of gene expression in response to water deficit of chickpea (Cicer arietinum L.) varieties differing in drought tolerance. BMC Plant Biol 10:24
Mantri NL, Ford R, Coram TE, Pang ECK (2010) Evidence of unique and shared responses to major biotic and abiotic stresses in chickpea. Environ Exp Bot 69:286–292
Coram TE, Pang ECK (2006) Expression profiling of chickpea genes differentially regulated during a resistance response to Ascochyta rabiei. Plant Biotechnol J 4:647–666
Coram TE, Pang ECK (2007) Transcriptional profiling of chickpea genes differentially regulated by salicylic acid, methyl jasmonate and aminocyclopropane carboxylic acid to reveal pathways of defence-related gene regulation. Funct Plant Biol 34:52–64
Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194
Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8:195–202
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35:W182–W185
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−△△CT method. Methods 25:402–408
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. Nat Genet 25:25–29
Weinl S, Kudla J (2009) The CBL-CIPK Ca2+-decoding signaling network: function and perspectives. New Phytol 184:517–528
Alamillo JM, Bartels D (1996) Light and stage of development influence the expression of desiccation-induced genes in the resurrection plant Craterostigma plantagineum. Plant Cell Environ 19:300–310
Ouvrard O, Cellier F, Ferrare K, Tousch D, Lamaze T, Dupuis JM, Casse-Delbart F (1996) Identification and expression of water stress- and abscisic acid-regulated genes in a drought-tolerant sunflower genotype. Plant Mol Biol 31:819–829
Zeng Q, Chen X, Wood AJ (2002) Two early light-inducible protein (ELIP) cDNAs from the resurrection plant Tortula ruralis are differentially expressed in response to desiccation, rehydration, salinity, and high light. J Exp Bot 53:1197–1205
Peng H, Cheng HY, Chen C, Yu XW, Yang JN, Gao WR, Shi QH, Zhang H, Li JG, Ma H (2009) A NAC transcription factor gene of Chickpea (Cicer arietinum), CarNAC3, is involved in drought stress response and various developmental processes. J Plant Physiol 166:1934–1945
Peng H, Yu X, Cheng H, Shi Q, Zhang H, Li JG, Ma H (2010) Cloning and characterization of a novel NAC family gene CarNAC1 from chickpea (Cicer arietinum L.). Mol Biotechnol 44:30–40
Coram TE, Pang ECK (2005) Isolation and analysis of candidate ascochyta blight defence genes in chickpea. Part I. Generation and analysis of an expressed sequence tag (EST) library. Physiol Mol Plant Pathol 66:192–200
Coram TE, Pang ECK (2005) Isolation and analysis of candidate ascochyta blight defence genes in chickpea. Part II. Microarray expression analysis of putative defence-related ESTs. Physiol Mol Plant Pathol 66:201–210
Ashraf N, Ghai D, Barman P, Basu S, Gangisetty N, Mandal MK, Chakraborty N, Datta A, Chakraborty S (2009) Comparative analyses of genotype dependent expressed sequence tags and stress-responsive transcriptome of chickpea wilt illustrate predicted and unexpected genes and novel regulators of plant immunity. BMC Genomics 10:415
Varshney RK, Hiremath PJ, Lekha P, Kashiwagi J, Balaji J, Deokar AA, Vadez V, Xiao Y, Srinivasan R, Gaur PM, Siddique KHM, Town CD, Hoisington DA (2009) A comprehensive resource of drought- and salinity-responsive ESTs for gene discovery and marker development in chickpea (Cicer arietinum L.). BMC Genomics 10:523
Tripathi V, Parasuraman B, Laxmi A, Chattopadhyay D (2009) CIPK6, a CBL-interacting protein kinase is required for development and salt tolerance in plants. Plant J 58:778–790
Matsumoto T, Wu J, Kanamori H, Katayose Y, Fujisawa M et al (2005) The Rice Annotation Project Database (RAP-DB): hub for Oryza sativa ssp. japonica genome information. Nature 436:500–793
Shinozaki K, Yamaguchi-Shinozaki K (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58:221–227
Seki M, Umezawa T, Urano K, Shinozaki K (2007) Regulatory metabolic networks in drought stress responses. Curr Opin Plant Biol 10:296–302
Lee BR, Jung WJ, Lee BH, Avice JC, Ourry A, Kim TH (2008) Kinetics of drought-induced pathogenesis-related proteins and its physiological significance in white clover leaves. Physiol Plant 132:329–337
Dunwell JM, Gibbings JG, Mahmood T, Naqvi SMS (2008) Germin and germin-like proteins: evolution, structure, and function. Crit Rev Plant Sci 27:342–375
Zhang Y, Xu WY, Li ZH, Deng XW, Wu WH, Xue YB (2008) F-box protein DOR functions as a novel inhibitory factor for abscisic acid-induced stomatal closure under drought stress in arabidopsis. Plant Physiol 148:2121–2133
Singh KB, Ocampo B, Robertson LD (1998) Diversity for abiotic and biotic stress resistance in the wild annual Cicer species. Genet Resour Crop Evol 45:9–17
Acknowledgments
We gratefully acknowledge the partial financial support from the projects supported by the National Natural Science Foundation of China (30960201, 30960206, 30860152 and 31160306), from the project supported by the Xinjiang Science and Technology Department of China (200991254), from the projects supported by the National Science and Technology Ministry (2006BAD09A04, 2006BAD09A08), from the project supported by the National Science Foundation for Postdoctoral Scientists of China (20080431107), from the project supported by the Jiangsu Science Foundation of Postdoctoral Scientists of China (0801048B) for this research.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Xiansheng Wang and Ying Liu contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, X., Liu, Y., Jia, Y. et al. Transcriptional responses to drought stress in root and leaf of chickpea seedling. Mol Biol Rep 39, 8147–8158 (2012). https://doi.org/10.1007/s11033-012-1662-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-1662-4