Abstract
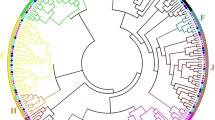
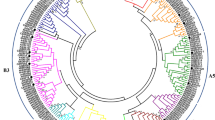
C2H2 proteins belong to a group of transcription factors (TFs) existing as a superfamily that plays important roles in defense responses and various other physiological processes in plants. The present study aimed to screen for and identify C2H2 proteins associated with defense responses to abiotic and biotic stresses in Carica papaya L. Data were collected for 47,483 papaya-expressed sequence tags (ESTs). The full-length cDNA nucleotide sequences of 87 C2H2 proteins were predicated by BioEdit. All 91 C2H2 proteins were aligned, and a phylogenetic tree was constructed using DNAman. The expression levels of 42 C2H2 were analyzed under conditions of salt stress by quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR). Methyl jasmonate treatment rapidly upregulated ZF 23.4 and ZF 30,912.1 by 18.6- and 21.7-fold, respectively. ZF 1.3, ZF 138.44, ZF 94.49, ZF29.160, and ZF 20.206 were found to be downregulated after low temperature treatment at very significant levels (p < 0.01). ZF 23.4, ZF 161.1, and ZF 30,912.1 were upregulated while ZF1.3, ZF 158.1, ZF 249.5, ZF 138.44, ZF 94.49, ZF 29.160, and ZF20.206 were significantly downregulated by Spermine treatment. ZF 23.4 was upregulated while ZF 1.3, ZF 249.5, ZF94.94, ZF 29.160, ZF 138.44, and ZF 20.206 were significantly repressed after SA treatment. ZF 23.4 and ZF 30,912.1 were significantly upregulated after sap inoculation with papaya ringspot virus pathogen. ZF 30,912.1 was subcellularly localized in the nucleus by a transgenic fusion of pBS–ZF 30,912.1–GFP into the protoplast of papaya. The results of the present study showed that ZF 30,912.1 could be an important TF that mediates responses to abiotic and biotic stresses in papaya.







Similar content being viewed by others
Abbreviations
- ABA:
-
Abscisic acid
- SA:
-
Salicylic acid
- C2H2:
-
Cys2/His2 finger protein
- EAR:
-
ERF associated amphiphilic repression
- GFP:
-
Green fluorescent protein
- MeJA:
-
Methyl-jasmonate
- PEG:
-
Polyethylene glycol
- RNAi:
-
RNA interference
- Spm:
-
Spermine
- TF:
-
Transcription factor
- TFPs:
-
Transcription factor proteins
- VIGS:
-
Virus induced gene silencing
- ZF:
-
Zinc finger
- ZFP:
-
Zinc finger protein
- qRT-PCR:
-
Real-time quantitative PCR
References
Zhang James Z, Creelman Robert A, Zhu Jian-Kang (2004) From laboratory to field. Using information from Arabidopsis to engineer salt, cold, and drought tolerance in crops. Plant Physiol 135:615–621
Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9:436–442
Huang J, Yang X, Wang MM, Tang HJ, Ding LY, Shen Y, Zhang HS (2007) A novel rice C2H2-type zinc finger protein lacking DLN-box/EAR-motif plays a role in salt tolerance. Biochim Biophys Acta 1769:220–227
Tian ZD, Zhang Y, Liu J, Xie CH (2010) Novel potato C2H2-type zinc finger protein gene, StZFP1, which responds to biotic and abiotic stress, plays a role in salt tolerance. Plant Biol (Stuttg) 12(5):689–697
Kubo K, Sakamoto A, Kobayashi A, Rybka Z, Kanno Y, Nakagawa H, Takatsuji H (1998) Cys2/His2 zinc-finger protein family of petunia: evolution and general mechanism of target-sequence recognition. Nucleic Acids Res 26:608–615
Englbrcht CC, Schoof H, Bohm S (2004) Conservation diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics 5:39
Agarwal P, Arora R, Ray S, Singh AK, Singh VP, Takatsuji H, Kapoor S, Tyagi AK (2007) Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol Biol 65:467–485
Sun SJ, Guo SQ, Yang X, Bao YM, Tang HJ, Sun H, Huang J, Zhang HS (2010) Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. J Exp Bot 61(10):2807–2818
Ming R, Hou S, Feng Y et al (2008) The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452:991–996
Xu SM, Wang XC, Chen J (2007) Zinc finger protein 1 (ThZF1) from salt cress (Thellungiella halophila) is a Cys-2/His-2-type transcription factor involved in drought and salt stress. Plant Cell Rep 26:497–506
He XJ, Mu RL, Cao WH, Zhang ZG, Zhang JS, Chen SY (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J 44:903–916
Cifrci-Yilmaz S, Morsy MR, Song L, Coutu A, Krizek BA, Lewis MW, Warren D, Cushman J, Connolly EL, Mittler R (2007) The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J Biol Chem 282:9260–9268
Dai X, Xu Y, Ma Q, Xu W, Wang T, Xue Y, Chong K (2007) Overexpression of a R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol 143:1739–1751
Huang XY, Chao DY, Gao JP, Zhu MZ, Shi M, Lin HX (2009) A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev 23:1805–1817
Oh S, Song S, Kim Y, Jang H, Kim S, Kim M, Kim Y, Nahm B, Kim J (2005) Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol 138:341–351
Li G, Tai FJ, Zheng Y, Luo J, Gong SY, Zhang ZT, Li XB (2009) Increased tolerance of rice to cold, drought and oxidative stresses mediated by the overexpression of a gene that encodes the zinc finger protein ZFP245. Biochem Biophys Res Commun 389(3):556–561
Cuevas JC, Lopez-Cobollo R, Alcazar R, Zarza X, Koncz C, Altabell T, Salinas J, Tiburcio AF, Ferrando A (2009) Putrescine as a signal to modulate the indispensable ABA increase under cold stress. Plant Signal Behav 4:219–220
Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol 47:141–153
Vogel JT, Zarka DG, Buskirk VHA, Fowler SG, Thomashow MF (2005) Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 41(2):195–211
Kim JC, Lee SH, Cheng YH, Yoo CM, Lee SI, Chun HJ, Yun DJ, Hong JC, Lee SY, Lim CO, Cho MJ (2001) A novel cold inducible zinc finger protein from soybean, SCOF-1, enhances cold tolerance in transgenic plant. Plant J 25:247–259
Li G, Tai FJ, Zheng Y, Luo J, Gong SY, Zhang ZT, Li XB (2010) Two cotton Cys2/His2-type zinc-finger proteins, GhDi19-1 and GhDi19-2, are involved in plant response to salt/drought stress and abscisic acid signaling. Plant Mol Biol 74:437–452
Lorenzo O, Solano R (2005) Molecular players regulating the jasmonate signalling network. Curr Opin Plant Biol 8:532–540
Lorenzo O, Chico JM, Sanchez-Serrano JJ, Solano R (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16:1938–1950
Koornneef A, Pieterse CM (2008) Cross talk in defense signaling. Plant Physiol 146:839–844
Wang Z, Cao GY, Wang XL, Miao J, Liu XT, Chen ZL, Qu LJ, Gu HY (2008) Identification and characterization of COI1-dependent transcription factor genes involved in JA-mediated response to wounding in Arabidopsis plants. Plant Cell Rep 27:125–135
Yang B, Jiang YQ, Rahman MH, Deyholos MK, Kav NN (2009) Identification and expression analysis of WRKY transcription factor genes in canola (Brassica napus L.) in response to fungal pathogens and hormone treatments. BMC Plant Biol 9:68
Groppa MD, Benavides MP (2008) Polyamines and abiotic stress: recent advances. Amino Acids 34:35–45
Alonso-Blanco C, Aarts MGM, Bentsink L, Keurentjes JJB, Reymond M, Vreugdenhil D, Koornnee M (2009) What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 21(7):1877–1896. www.plantcell.org/cgi/doi/10.1105/tpc.109.068114
Gill SS, Tuteja N (2010) Polyamines and abiotic stress tolerance in plants. Plant Signal Behav 7:5(1)
Kam J, Gresshoff PM, Shorter R, Xue GP (2008) The Q-type C2H2 zinc finger subfamily of transcription factors in Triticum aestivum is predominantly expressed in roots and enriched with members containing an EAR repressor motif and responsive to drought stress. Plant Mol Biol 67:305–322
Wang GF, Wang H, Zhu J, Zhang J, Zhang XW, Wang F, Tang YP, Mei B, Xu ZK, Song RT (2010) An expression analysis of 57 transcription factors derived from ESTs of developing seeds in Maize (Zea mays). Plant Cell Rep 29:545–559
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real time quantitative PCR and the 2−∆∆Ct method. Methods 25:402–408
Yoo Sang-Dong, Cho Young-Hee, Sheen Jen (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2(7):1565–1572
Jiang L, Wang J, Liu Z, Wang L, Zhang F, Liu GC, Zhong Q (2010) Silencing induced by inverted repeat constructs in protoplasts of Nicotiana benthamiana. Plant cell tissue organ cult 100:139–148
Tian LM, Huang CL, Zhang XH, Zhang LS, Wu ZY (2005) Advances of plant zinc finger proteins involved in abiotic stress. Biotechnol Bull 6:13–16
Chen WQ, Provart NJ, Glazebrook J, Katagiri F, Chang HS, Eulgem T, Mauch F, Luan S, Zou GZ, Whitham SA, Budworth PR, Tao Y, Xie ZY, Chen X, Lam S, Kreps JA, Harper JF, Si-Ammour A, Mauch-Mani B, Heinlein M, Kobayashi K, Hohn T, Dang JL, Wang X, Zhua T (2002) Expression profile matrix of arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14:559–574
Huang J, Wang JF, Zhang HS (2004) Structure and function of plant C2H2 zinc finger protein. Hereditas (Beijing) 26(3):414–418
Miller J, Mclachlan AD, Klug A (1985) Repetitive zinc-binding domains in the protein transcription factor 111A from xenopus oocytes. EMBOJ 4:1609–1614
Nakashima K, Ito Y, Yamaguchi-Shinozaki K (2009) Transcriptional regulatory networks in response to abiotic stresses in arabidopsis and grasses. Plant Physiol 149:88–95
Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low temperature, or high-salt stress. Plant Cell 6:251–264
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Dong XN (1996) SA, JA, ethylene, and disease resistance in plants. Curr Opin Plant Biol 1:316–323
Lin R, Zhao W, Meng X, Wang M, Peng Y (2007) Rice gene OsNAC19 encodes a novel NAC-domain transcription factor and responds to infection by Magnaporthe grisea. Plant Sci 172:120–130. doi:101016/j.plantsci.2006.07.019
Acknowledgment
The project was supported by the National Natural Science Foundation of China (grants No. 30771477 and No. 30571277).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.






11033_2012_1542_MOESM6_ESM.jpg
Supplementary Data Fig. 2 (a) Nucleotide sequence of ZF30912.1 and its amino acid sequence. (b) Comparison of the amino acid sequence of ZF30912.1. The gene sequence of ZF 30912.1 was compared with 12 ZF with known functions; their accession numbers are as follows: Capsicum annuu CAZFP1 (AT196704.1), Glycine max SCOF1 (GMU68763), Petunia ZPT2-1 (X60700), Arabidopsis thaliana AZF1 (NM126145), Brassica rapa BCZFP1 (AAB53261), Petunia × hybrida ZPT3-3 (AB035133), Petunia × hybrida ZPT2-10 (BAA96070), Arabidopsis thaliana AZF3 (NM-123683), Arabidopsis thaliana AZF1 (NM-001125191), Petunia ZPT2-2 (D26038), Oryza sativa Japanica Group OSZFP34 (AAPA2273), Silene latifolia ZPT3-2 (DQ017765), and Carica papaya ZF30912.1 (gb|ABIM01030863.1|). (JPEG 182 kb)

Rights and permissions
About this article
Cite this article
Jiang, L., Pan, Lj. Identification and expression of C2H2 transcription factor genes in Carica papaya under abiotic and biotic stresses. Mol Biol Rep 39, 7105–7115 (2012). https://doi.org/10.1007/s11033-012-1542-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-1542-y