Abstract
Sulfite oxidase (SO) catalyzes the oxidation of sulfite to sulfate and thus has important roles in diverse metabolic processes. However, systematic molecular and functional investigations on the putative SO from tobacco (Nicotiana benthamiana) have hitherto not been reported. In this work, a full-length cDNA encoding putative sulfite oxidase from N. benthamiana (NbSO) was isolated. The deduced NbSO protein shares high homology and typical structural features with other species SOs. Phylogenetic analysis indicates that NbSO cDNA clone encodes a tobacco SO isoform. Southern blot analysis suggests that NbSO is a single-copy gene in the N. benthamiana genome. The NbSO transcript levels were higher in aerial tissues and were up-regulated in N. benthamiana during sulfite stress. Reducing the SO expression levels through virus-induced gene silencing caused a substantial accumulation in sulfite content and less sulfate accumulation in N. benthamiana leaves when exposed to sulfite stress, and thus resulted in decreased tolerance to sulfite stress. Taken together, this study improves our understanding on the molecular and functional properties of plant SO and provides genetic evidence on the involvement of SO in sulfite detoxification in a sulfite-oxidizing manner in N. benthamiana plants.





Similar content being viewed by others
References
Hille R (1996) The mononuclear molybdenum enzymes. Chem Rev 96:2757–2816
Kisker C, Schindelin H, Pacheco A, Wehbi WA, Garrett RM, Rajagopalan KV, Enemark JH, Rees DC (1997) Molecular basis of sulfite oxidase deficiency from the structure of sulfite oxidase. Cell 91:973–983
Mendel RR, Schwarz G (1999) Molybdoenzymes and molybdenum cofactor in plants. Crit Rev Plant Sci 18:33–69
Cohen HJ, Fridovich I (1971) Hepatic sulfite oxidase. The nature and function of the heme prosthetic groups. J Biol Chem 246:367–373
Cohen HJ, Fridovich I (1971) Hepatic sulfite oxidase. Purification and properties. J Biol Chem 246:359–366
Kessler DL, Rajagopalan KV (1972) Purification and properties of sulfite oxidase from chicken liver. J Biol Chem 247:6566–6573
Garrett RM, Rajagopalan KV (1994) Molecular cloning of rat liver sulfite oxidase. J Biol Chem 269:272–276
Cohen HJ, Betcher-Lange S, Kessler DL, Rajagopalan KV (1972) Hepatic sulfite oxidase. Congruency in mitochondria of prosthetic groups and activity. J Biol Chem 247:7759–7766
Kisker C, Schindelin H, Rees DC (1997) Molybdenum cofactor-containing enzymes: structure and mechanism. Annu Rev Biochem 66:233–267
Brody MS, Hille R (1999) The kinetic behavior of chicken liver sulfite oxidase. Biochemistry 38:6668–6677
Garrett RM, Johnson JL, Graf TN, Feigenbaum A, Rajagopalan KV (1998) Human sulfite oxidase R160Q: identification of the mutation in a sulfite oxidase-deficient patient and expression and characterization of the mutant enzyme. Proc Natl Acad Sci USA 95:6394–6398
Eilers T, Schwarz G, Brinkmann H, Witt C, Richter T, Nieder J, Koch B, Hille R, Hansch R, Mendel RR (2001) Identification and biochemical characterization of Arabidopsis thaliana sulfite oxidase- a new player in plant sulfur metabolism. J Biol Chem 276:46989–46994
Schrader N, Fisher K, Theis K, Mendel RR, Schwarz G, Kisker C (2003) The crystal structure of plant sulfite oxidase provides insights into sulfite oxidation in plants and animals. Structure 11:1251–1263
Hansch R, Lang C, Riebeseel E, Lindigkeit R, Gessler A, Rennenberg H, Mendel RR (2006) Plant sulfite oxidase as novel producer of H2O2: combination of enzyme catalysis with a subsequent non-enzymatic reaction step. J Biol Chem 281:6884–6888
Byrne RS, Hänsch R, Mendel RR, Hille R (2009) Oxidative half-reaction of Arabidopsis thaliana sulfite oxidase: generation of superoxide by a peroxisomal enzyme. J Biol Chem 284:35479–35484
Nowak K, Luniak N, Witt C, Wustefeld Y, Wachter A, Mendel RR, Hansch R (2004) Peroxisomal localization of sulfite oxidase separates it from chloroplast-based sulfur assimilation. Plant Cell Physiol 45:1889–1894
Brychkova G, Xia Z, Yang G, Yesbergenova Z, Zhang Z, Davydov O, Fluhr R, Sagi M (2007) Sulfite oxidase protects plants against sulfur dioxide toxicity. Plant J 50:696–709
Lang C, Popko J, Wirtz M, Hell R, Herschbach C, Kreuzwieser J, Rennenberg H, Mendel RR, Hansch R (2007) Sulphite oxidase as key enzyme for protecting plants against sulphur dioxide. Plant Cell Environ 30:447–455
Goodin MM, Zaitlin D, Naidu RA, Lommel SA (2008) Nicotiana benthamiana: its history and future as a model for plant-pathogen interactions. Mol Plant Microbe Interact 21:1015–1026
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Asai S, Ohta K, Yoshioka H (2008) MAPK signaling regulates nitric oxide and NADPH oxidase-dependent oxidative bursts in Nicotiana benthamiana. Plant Cell 20:1390–1406
Constantin GD, Krath BN, MacFarlane SA, Nicolaisen M, Johansen IE, Lund OS (2004) Virus-induced gene silencing as a tool for functional genomics in a legume species. Plant J 40:622–631
Qian W, Yu C, Qian H, Liu X, Zhang A, Johansen IE, Wang D (2007) Molecular and functional analysis of phosphomannomutase (PMM) from higher plants and genetic evidence for the involvement of PMM in ascorbic acid biosynthesis in Arabidopsis and Nicotiana benthamiana. Plant J 49:399–413
Liu Y, Nakayama N, Schiff M, Litt A, Irish VF, Dinesh-Kumar SP (2004) Virus induced gene silencing of a DEFICIENS ortholog in Nicotiana benthamiana. Plant Mol Biol 54:701–711
MacFarlane SA, Popovich AH (2000) Efficient expression of foreign proteins in roots from tobravirus vectors. Virology 267:29–35
Nakamura T, Meyer C, Sano H (2002) Molecular cloning and characterization of plant genes encoding novel peroxisomal molybdoenzymes of the sulphite oxidase family. J Exp Bot 53:1833–1836
Hansch R, Lang C, Rennenberg H, Mendel RR (2007) Significance of plant sulfite oxidase. Plant Biol (Stuttg) 9:589–595
Lindboet JA, Fitzmaurice WP, della-Cioppa G (2001) Virus mediated reprogramming of gene expression in plants. Curr Opin Plant Biol 4:181–185
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (30971548) and The Key Project of Science and Technology of Henan Tobacco Company (HYKJ201010). The authors thank Dr Daowen Wang (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China) for generously providing the PEBV-based pCAPE vector system.
Author information
Authors and Affiliations
Corresponding author
Additional information
Zongliang Xia and Xinhong Su contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2011_993_MOESM1_ESM.pdf
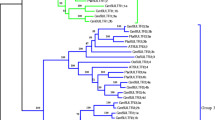
Fig. 1 Sequence alignment and phylogenetic analysis of sulfite oxidase (SO) from N. benthamiana and other species (A) An alignment is shown for the deduced amino acid sequence of SO from Nicotiana benthamiana (NbSO), Lycopersicon esculentum (LeSO), Solanum tuberosum (StSO), Arabidopsis thaliana (AtSO), Oryza sativa (OsSO), and Zea Mays (ZmSO). The numbers on the left indicate the amino acid position. Identical residues in all these proteins are shown in a black background. Dashes indicated gaps introduced for optimal alignment. The putative molybdopterin-binding domain and dimerization domain are underlined with a solid line and a dashed line, respectively. The boxed regions mark the predicted signal peptide. (B) Phylogenetic tree based on SO protein sequences from bacterial, plants, and animals. The bootstrap values shown were calculated based on 500 replications. The tree was constructed using the neighbor-joining method. NbSO, Nicotiana benthamiana (HQ699883); LeSO, Lycopersicon esculentum (DQ853413); StSO, Solanum tuberosum (DQ284487); AtSO, Arabidopsis thaliana (At3g01910); BoSO, Brassica oleracea (AC183495); OsSO, Oryza sativa (Os08g0530400); ZmSO, Zea Mays (FJ436404); PtSO, Populus trichocarpa (XP_002300104); HcSO, Hibiscus cannabinus (ACU33027); DmSO, Drosophila melanogaster (NM_133103); CeSO, Caenorhabditis elegans (NM_001029564); GgSO, Gallus gallus (P07850); HsSO, Homo sapiens (NM_000456); MmSO, Mus musculus (NM_173733); RnSO, Rattus norvegicus (NM_031127); EcSO, Escherichia coli (NP_288430). Fig. 2 Southern blot analysis of NbSO The genomic DNA samples from N. benthamiana were digested separately with either EcoRI, XbaI or BamHI. The hybridization bands are indicated by arrows. DNA size markers (HindIII digest of lambda DNA) are shown on the right. The results shown are typical of three independent hybridizations. (PDF 91 kb)
Rights and permissions
About this article
Cite this article
Xia, Z., Su, X., Wu, J. et al. Molecular cloning and functional characterization of a putative sulfite oxidase (SO) ortholog from Nicotiana benthamiana . Mol Biol Rep 39, 2429–2437 (2012). https://doi.org/10.1007/s11033-011-0993-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-0993-x