Abstract
As soybean plays an indispensable role in the supply of vegetable oil and protein, balancing the relationship between seed quality and yield traits according to human demand has become an important breeding goal for soybean improvement. Here, 256 intraspecific recombinant inbred lines (RILs), derived from a cross between Qi Huang No.34 (QH34) and Ji Dou No.17 (JD17), were used for quantitative trait loci (QTLs) mapping with remarkable four chemical and physical properties with a purpose for exploring the distribution of excellent alleles in germplasm resources in China. A total of 25 QTLs were detected, of which 10 QTLs inherited the alleles from the parent QH34. Pedigree research on favorable alleles on these QTLs showed the process of excellent alleles pyramided into QH34. Meta-analysis of the 25 QTLs by comparing with existed QTLs in previous study identified 17 novel QTLs. QTLs with pleiotropic effects have been detected. Furthermore, three representative elite recombinant inbred lines in different locations that have great potential in soybean breeding were selected, and finally, four seed weight-related candidate genes were identified. The discovery of these QTLs provides a new guidance for combining the diversity and rarity of germplasm resources, which can effectively increase population genetic diversity and broaden genetic basis of varieties.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soybean [Glycine max (L.) Merr.] is an important food and feed crop, and a major source of feed protein and vegetable oil, which is owning to its chemical and physical properties of seed protein, oil content, and 100-seed weight (Yang et al. 2019). Approximately, the protein and oil contents in dry soybean seeds are 40% and 20%, respectively (Rajcan et al. 2005). Seed weight is a component of soybean yield and is an indicator of seed size and seed plumpness. These are complex quantitative genetic traits that are controlled by multiple genes and affected by environmental conditions.
Many studies that focused on QTL mapping to identify QTLs for seed protein content, oil content, seed protein plus oil content, and 100-seed weight in soybean (Chapman et al. 2003; Diers et al. 1992; Orf et al. 1999; Pathan et al. 2013; Sebolt et al. 2000; Li et al. 2018; Yan et al. 2017). To date, 248 QTLs for protein content, 327 for soybean oil content, five for protein plus oil content, and 304 for soybean 100-seed weight distributed across 20 pairs of chromosomes have been reported in SoyBase (http://www.soybase.org). Although numerous seed quality and yield-related QTLs have been detected, only two genes have been cloned, GmOLEO1 enhanced oil accumulation by affecting triacylglycerol metabolism (Zhang et al. 2019). GmSWEET39 is positively correlated with soybean seed oil content in terms of the expression level (Miao et al. 2020). Previous studies have shown that soybean seed oil content and 100-seed weight are positively correlated; however, seed protein content is negatively correlated with seed oil content and 100-seed weight, respectively (Wilcox and Zhang 1997; Wilcox 1998). These negative correlations make it difficult to improve those traits simultaneously by traditional breeding.
The rapid progress of soybean pan-genome research has made it possible to obtain pan-genetic sites that control classic traits such as seed protein content, oil content, seed protein plus oil content, and 100-seed weight. China, where cultivar soybean began, has the most abundant soybean germplasm resources and gene pool in the world. Liu et al. (2020) selected 26 accessions (20 of them originated from China), including Qi Huang No.34 and Ji Dou No.17, for de novo assembly to construct a well representative pan-genome, which is representative in terms of phylogenetic relationships and geographic distributions. A pedigree study revealed that QH34 inherited lineages from six out of these 26 accessions in China and the pedigree of JD17 comes from the USA, Japan, North Korea, and other places (Fig. 1). Besides different geographical origins and genetic backgrounds, QH34 and JD17 differ greatly in protein content, oil content, and 100-seed weight, which lead to a relatively wide range of distribution for their progenies; thereby, it is easier to select new varieties with elite traits with different environmental adaptability than improving or cultivating the parental lines.
Pedigree analysis of QH34 and JD17. a, b Pedigree diagram of QH34 and JD17, respectively. QH34 was genetically inherited the predominant alleles from the six elite accessions among the 26 germplasm, which were sequenced by Liu et al. (2020) and highlighted in blue
In this study, QTLs associated with soybean seed protein content, oil content, seed protein plus oil content, and 100-seed weight were detected using the RILs derived from a cross between QH34 and JD17. Pedigree research on favorable alleles on 10 QTLs which inherited the alleles from the parent QH34 showed the process of excellent alleles pyramided into QH34. Four candidate genes related to 100-seed weight were detected as the significantly differential expression between the two parents. The novel QTLs may provide new breeding ideas for improving the seed quality and yield, and guide soybean production and industrialization development.
Materials and methods
Plant materials and growth conditions
A population consisted of 256 F8:9 RILs was deployed for QTL mapping. The maternal line of the population, QH34, was derived from a cross between Youchu No.4 and 86573-16, and was developed by Shandong Academy of Agricultural Sciences (Institute of Crop Research). The other paternal line, JD17, was derived from a cross between Hobbit and Zao 5241. JD17 was released by Hebei Academy of Agricultural and Forestry Sciences (Institute of Grain and Oil Crops Research) in China.
The two parents and RILs were planted at three locations including Sanya (18.24 N, 109.50′ E), Nanjing (32.04 N, 118.76′ E), and Beijing (39.9 N, 116.24′ E) in 2019, with a row space of 0.5 m (ten seeds in a row) and 0.1 m between lines. The harvested seeds were used for trait evaluation.
Phenotype evaluation
The harvested seeds of two parents and RILs in Sanya, Nanjing, and Beijing were used for 100-seed weight measurement with an electronic scale (JY3002, Shanghai Sunny Hengping Scientific Instrument Co., Ltd.); the seed oil and seed protein content was analyzed on a Fourier transform near-infrared spectroscopy in Chinese Academy of Agricultural Sciences. All the experiments were performed with three replicates. Seed protein plus oil content was defined as the sum of protein content and oil content.
Constructing the genetic map
The parental lines, QH34 and JD17, as well as the 256 F6:7 RILs population, were sequenced with the reduced representation sequencing method of specific-locus amplified fragment sequencing (SLAF-seq) on an Illumina sequencing platform for developing SNP markers. Sequenced results were aligned to the Williams 82 reference genome (Wm82.a2.v1). A total of 2,223,219 SNPs were obtained in this project, and 1,276,728 SNPs were successfully typed. SNP filter and quality check yielded 6402 high confidence SNP markers that distributed over 20 linkage groups. The 6402 SNP markers were then used for constructing the genetic map using the HighMap software. The total length of the assembled genetic map was 1726.03 cM.
RNA-seq analysis
The parental lines, QH34 and JD17, were grown in Beijing in 2018. Self-pollinated and premature or mature seeds at stage I (E2 stage), stage II (M2 stage), and stage III (L1 stage) were collected, respectively, and the total RNA was extracted from the seeds tissues with a DP411 RNAprep pure Plant Kit TIANGEN. RNA samples with high quality and RIN (RNA intact number) values (RIN > 6) were used for sequencing on an Illumina platform with the parameter of pair-end 150 (PE150) (Annoroad Gene Technology (Beijing) Co., LTD, Hangzhou, China). Gene expression level was quantified with FPKM (Fragments per Kilobase per Million Mapped Fragments) as below formula:
If FPKM (A) is the expression level of gene A, then F is the number of fragments uniquely aligned to gene A, N is the total number of fragments uniquely aligned to the reference gene, and L is the length of the exon region of gene A.
The |log2 Fold change|≥1 with q-value (adjusted p-value) <0.05 was used for differential expressed gene analysis.
Statistical analysis
The mean value of three replicates for protein, oil, protein plus oil content, and 100-seed weight was used for correlation analysis and QTL mapping. Linear regression analysis (pearson correlation coefficient) of these four traits was analyzed with R statistical package and used to calculate the phenotypic correlation coefficients. Genetic linkage map of QH34 × JD17 was constructed using the F6:7 population with SNP markers generated from SLAF-seq technology. QTL mapping was conducted with the software QTL IciMapping V4.1 using the inclusive composite interval mapping (ICIM) model (Li et al. 2007). A LOD score > 2.0 was used to declare the existence of a QTL in a specific genomic region.
Results
Phenotypic analysis of quality and yield traits in Parents and RILs
Seed protein content, seed oil content, seed protein plus oil content and 100-seed weight measured in this study were selected to represent soybean quality and yield traits that differ between QH34 and JD17. QH34 is a typical high-protein inbred line with high yield; the protein content shows great variance planting from south to north of China, which is 39.93%, 42.53%, 44.99% respectively in Sanya, Nanjing, and Beijing. The oil content of QH34 is 21.58%, 21.81%, and 20.22% respectively from south to north; seed protein plus oil content shows an increase from south to north, which is 61.51%, 64.34%, and 65.21% respectively. And the 100-seed weight shows obvious decrease when grown in Nanjing (22.62 g); no significant difference in Sanya and Beijing, 27.57 g and 27.80 g, respectively (Fig. 2). While in JD17, a typical high-oil inbred, the yield is lower than QH34. The protein content of JD17 is 39.74%, 38.51%, and 42.45% respectively from south to north, and the protein content is significantly lower than QH34 in Nanjing and Beijing (P < 0.01), while no significant difference when grown in Sanya (Fig. 2a). The oil content shows a highest level of 24.03% in Nanjing, then Sanya and Beijing (22.39% and 21.20%, respectively); Except in Beijing, the oil content of QH34 and JD17 shows extremely significant difference in both Sanya and Nanjing (P < 0.01) (Fig. 2b). The seed protein plus oil content of JD17 shows a similar increase from south to north, which is 62.13%, 62.54%, and 63.65%, respectively, in above three locations, and the protein plus oil content is significantly lower than the QH34 in Nanjing and Beijing (Fig. 2c). In terms of 100-seed weight, similar with QH34, JD17 shows a highest level in Beijing (21.35 g) then Sanya (19.82 g) and Nanjing (18.13 g), while it is significantly lower than that was in QH34 (Fig. 2d).
Phenotypes of QH34 and JD17 planted in Sanya, Nanjing and Beijing. a The protein content of QH34 and JD17 planted in Sanya, Nanjing, and Beijing. b The oil content of QH34 and JD17 planted in Sanya, Nanjing, and Beijing. c The protein plus oil content of QH34 and JD17 planted in Sanya, Nanjing, and Beijing. d The 100-seed weight planted in Sanya, Nanjing, and Beijing
The phenotypic variation of the RILs at three different locations involving three soybean seed quality and one yield-related traits was documented in Table 1. All four soybean seed–related traits showed continuous variation and approximately in accordance with a normal distribution, with absolute vales of both skewness and kurtosis being less than 1.0, with the exception of kurtosis for 100-seed weight in Sanya (SHW) as well as protein plus oil content and 100-seed weight in Beijing (BPplusO and BHW) (Table 1). Notably, the phenotypic data of all four traits exhibited obvious bidirectional transgressive segregation in this population, indicating the polygenic control of the soybean seed quality and yield traits (Fig. 3). The Pearson correlation analysis for phenotypic values of the RIL population in each location was shown in Table 2. It is obvious that the 100-seed weight is positively correlated with soybean protein content and soybean protein plus oil content, with a significance of P < 0.001 in Nanjing and Beijing, and P < 0.05 in Sanya. There is no significant (P > 0.05) correlation with between 100-seed weight and seed oil content. Seed protein plus oil content has a very significant (P < 0.001) positive correlation with the protein content, and a very significant (P < 0.001) negative correlation with the oil content in each location. An extremely high negative correlation existed between seed protein content and seed oil content in each location (P < 0.001) (Table 2).
QTL mapping of seed quality and yield traits in the RIL population
A total of 25 significant QTLs across 14 chromosomes (including chromosomes 1, 3, 4, 5, 6, 7, 8, 9, 11, 13, 17, 18, 19, and 20) were identified with the explained phenotypic variance of 3.07–11.24% and the LOD values of 2.07–6.93 (Table 3; Fig. 4). These 25 QTLs corresponded to the four soybean seed quality and yield traits in different locations, with three for protein content, ten for oil content, three for seed protein plus oil content, and nine for 100-seed weight. Among that, Chr 11 contains most QTLs involving 100-seed weight, protein plus oil content in Nanjing, and protein content in Sanya. Chr 4 contains three independent QTLs involving 100-seed weight in three locations. Chr 5 also contains three tandem QTLs involving oil and protein content in Nanjing, and protein plus oil content in Beijing (Table 3). The highest explained phenotypic variance and LOD value were found for a 100-seed weight QTL (qNHW11-1). Ten QTLs with a negative additive effect inherited the contributable alleles from the parent QH34, while 15 with a positive additive effect inherited the contributable alleles from the parent JD17 (Table 3).
QTL distribution for seed quality and yield-related trait detected from the RIL population across three different locations. a The QTL distribution for seed protein content. b The QTL distribution for seed oil content. c The QTL distribution for seed protein plus oil content. d The QTL distribution for 100-seed weight. Different colored lines within the chromosome cycle represent the chromosome positions and LOD values for QTLs detected for protein content in Sanya (SP), protein content in Nanjing (NP), and protein content in Beijing (BP); oil content in Sanya (SO), oil content in Nanjing (NO), and oil content in Beijing (BO); protein plus oil content in Sanya (SPplusO), protein plus oil content in Nanjing (NPplusO), and protein plus oil content in Beijing (BPplusO); 100-seed in Sanya (SHW), 100-seed weight in Nanjing (NHW), and 100-seed weight in Beijing (BHW). Detailed QTL information is listed in Table 3
QTLs for protein content
In this study, a total of three QTLs related to seed protein content were identified with the explained phenotypic variance of 3.07–7.50% in two locations at Sanya (qSP11-1) and Nanjing (qNP5-1 and qNP3-1). QTL qNP5-1 (31.6–32.1 Mb) has the highest LOD value of 4.88 and explained 7.50% of the phenotypic variation. The contributable alleles for the protein content–related two QTLs (qNP3-1 and qSP11-1) from this study were inherited from the high-protein parent QH34, while the contribution of qNP5-1 was inherited from the low protein parent JD17 (Table 3; Fig. 4a). No confidential QTL-controlling soybean seed protein content was detected in Beijing.
QTLs for oil content
Compared to the protein content, oil content is relatively stable and insensitive to the environmental change. Here, totally ten soybean seed oil content–related QTLs were identified in Sanya (qSO8-1, qSO13-1, and qSO17-1), Nanjing (qNO5-1, qNO6-1, and qNO13-1), and Beijing (qBO1-1, qBO3-1, qBO8-1, and qBO19-1), which explained phenotypic variation of 3.27–9.38%. All three oil content–related QTLs detected in Sanya and four detected in Beijing were inherited from the high-oil parent JD17, except two QTLs (qNO5-1 and qNO6-1) identified in Nanjing, which were inherited from the low oil parent QH34. More interestingly, qNO6-1 explained the highest phenotypic variance of 9.38%, with the LOD value of 4.46. qSO8-1 on Chr 8 (46.3–46.5 Mb) has the highest LOD value of 5.16 and explained 7.44% of the phenotypic variation (Table 3; Fig. 4b).
QTLs for protein plus oil content
For the protein plus oil content trait, QTLs were detected in each of Sanya (qSPplusO9-1), Nanjing (qNPplusO11-1), and Beijing (qBPplusO5-1) with the LOD value around 2.5, which explained phenotypic variance from 4.22–6.28%. QTLs detected in both Sanya and Beijing were inherited from the parent JD17, while the QTL on Chr 11 detected in Nanjing was inherited from the parent QH34. The QTL qNPplusO11-1 on Chr 11 (2.37–2.63 Mb) detected in Nanjing explained the highest phenotype variation of 6.27%, and the contributable allele was inherited from large-high-protein seed parent QH34, indicating that the protein plus oil content is mostly related to the seed protein content and 100-seed weight. Interestingly, the other two QTLs of qSPplusO9-1 (Chr 9:42.1–42.2 Mb) and qBPplusO5-1 (Chr 5:33.57–33.65 Mb) detected in Sanya and Beijing, respectively, were inherited from the small-high-oil seed parent JD17 (Table 3; Fig. 4c).
QTLs for 100-seed weight
Nine QTLs related to 100-seed weight of soybean were identified, including two QTLs (qSHW4-1 and qSHW18-1) detected in Sanya, six QTLs (qNHW4-1, qNHW7-1, qNHW11-1, qNHW11-2, qNHW18-1, and qNHW20-1) detected in Nanjing and one QTL (qBHW4-1) detected in Beijing. QTL qNHW11-1 explained the highest phenotypic variance of 11.24% to 100-seed weight when grown in Nanjing, followed by qBHW4-1, explained a relatively high phenotypic variance of 7.44% to 100-seed weight when grown in Beijing. The contribution of most of the 100-seed weight-related QTLs were inherited from the high weight parent QH34 while only four QTLs (qNHW18-1, qNHW20-1, qNHW7-1, and qSHW18-1) were inherited from the low weight parent JD17 (Table 3; Fig. 4d).
Pedigree research on QTL favorable alleles
QH34 is a representative decent variety in China with wide adaptability and high yield and quality, which is proved by QH34-derived lineages from six representative accessions: 58–161, Xu Dou No.1, Tong Shan Tian E Dan, Feng Di Huang, Tie Jia Si Li Huang, and Ju xuan No.23. A pedigree research showed that six accessions above had different ways to contribute to the QTL favorable alleles on the 10 QTL-inherited alleles from the parent QH34.
Some alleles on QTLs were contributed by a single accession. For example, the alleles on qSHW4-1, increased 100-seed weight, were derived only from Ju Xuan No.23, and the alleles were not existed in other five accessions. Some accessions can provide complete alleles on QTL, for example, the alleles on qNHW4-1 were provided by 58–161, Xu Dou No.1, Tong Shan Tian E Dan, Feng Di Huang, and Ju xuan No.23 (any of them can provide complete qNHW4-1 favorable alleles individually). For another example, Xu Dou No.1 contained the whole favorable alleles on qBHW4-1. Some QTL favorable alleles were provided by different accessions, for example, the alleles on qNHW11-1 partly derived from 58 to 161 and Xu Dou No.1, and partly derived from Ju Xuan No.23 (Table S1; Fig. 5).
Pedigree research on QTL favorable alleles inherit from QH34. The alleles contributed by QH34 on different QTLs in six accessions are denoted as colored square. The accession contributed whole QTL favorable alleles and half of the alleles are denoted by square and half a square (rectangle), respectively. The red ellipse and arrow means the path of the QTL favorable alleles pyramided into QH34, and the gray ellipse means the unstudied accessions
The alleles on QTLs that exist or partially exist in multiple accessions occurred alleles pyramiding, for example, the QTL favorable alleles in 58–161 and Xu Dou No.1 pyramided in Ke Xi No.8, and QTL favorable alleles in both Tie Jia Si Li Huang and Feng Di Huang pyramided in Tie 4117. With the continuous hybridization, the QTL favorable alleles pyramided again until they pyramided into QH34. QTL favorable alleles pyramiding may make it difficult to lose the excellent genetic sites that affected the development of important plant traits. Interestingly enough, QTL favorable alleles on qSP11-1 were derived from Tong Shan Tian E Dan, but were partly from Xu Dou No.1, indicating that the QTL favorable alleles would be partially lost during the gene transfer process (Fig. 5).
Discussion
Identification of novel and pleiotropic QTLs
SoyBase (http://www.soybase.org) is the soybean database that includes all the QTLs or genes that have been identified so far. We further compared our QTL mapping results with the related QTLs in soybase. A total of eight QTLs shared the same or overlapping confidence intervals with the QTLs identified in previous studies, including one for seed protein content (Mao et al. 2013; Pathan et al. 2013), two for seed oil content (Diers et al. 1992; Lee et al. 1996), and five for 100-seed weight (Hacisalihoglu et al. 2018; Han et al. 2012; Hyten et al. 2004; Mian et al. 1996; Yang et al. 2011). More importantly, we found 17 novel QTLs controlling seed quality and yield-related traits (Fig. 6; Table 3).
QTLs with pleiotropic effects for seed protein content and oil content have been reported in previous studies (Brummer et al. 1997; Hyten et al. 2004; Mansur et al. 1996; Orf et al. 1999; Sebolt et al. 2000). Most of the studies showed that the oil and protein paradoxically existed in soybean, thus it is reasonable that one QTL has a positive effect on oil or protein while having a negative effect on the other (Wilcox 1998). However, there are few studies of pleiotropic effects on both seed weight and seed protein plus oil content. In this study, one QTL located on Chr 5 between markers M5C31683037 and M5C32119045 identified in Nanjing was associated with both seed protein content (qNP5-1) and seed oil content (qNO5-1). One QTL located on Chr 11 between markers M11C2375493 and M11C2638141 can regulate both seed protein plus oil content (qNPplusO11-1) and 100-seed weight (qNHW11-2). These results indicated these QTLs associated with four traits may have pleiotropic effects.
The relationship between environment and phenotype
The change trend of the 100-seed weight of the parents showed that genotype contributes more to 100-seed weight than environment. But for the same genotype, 100-seed weight will change along with change of the latitude, both QH34 and JD17 have the highest 100-seed weight in Beijing. The variation is not significant in protein content between QH34 and JD17 in Sanya, which demonstrated that the protein content is easier affected by environmental than the genetic factor. The pattern of protein content is similar to the 100-seed weight in JD17, while the protein content experienced a significant increase from south to north in QH34. Both QH34 and JD17 have higher protein content in Beijing than the other two locations. Although QH34 have greater protein content than JD17 in general, it is unwise to grow QH34 in Sanya because there is no significant advantage over JD17 once grow them in Sanya. For both QH34 and JD17, the oil content is highest in Nanjing; it shows an increase and followed by a decrease from south to north, indicating that it is more beneficial for the accumulation of oil in Nanjing. However, the high oil potential of JD17 cannot be released in Beijing. The changes of protein plus oil content were generally consistent with the trend of protein from south to north, indicating the contribution of “protein plus oil content” mainly comes from “protein” rather than “oil,” which is also supported by the fact that the protein content (~40%) takes up more of seed weight than the oil content (~20%), and the seed weight is often relies on the change of protein content.
Correlation among seed quality and yield traits
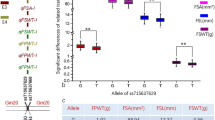
A highly significant negative correlation existed between seed protein and oil content in our study, which was in accordance with previous research (Wilcox 1998), indicating that it is difficult to increase both the oil and the protein content in soybean. It has been reported that there are large variations of correlation between seed weight and seed quality (seed protein and oil content), including positive and negative effects (Reinprecht et al. 2006; Specht et al. 2001). Here, the 100-seed weight was positively correlated with soybean protein, while no significant correlation with the oil content. However, some studies proved that soybean yield is negatively correlated with protein content but positively correlated with oil content (Wilcox and Zhang 1997). As we know, protein takes up more weight component than that is oil in soybean seed (~40% vs. 20%); 100-seed weight is only one of the component that contribute to the yield, and it is possible other yield factors also play the roles, i.e., by reducing the grain number per plant, resulting in a decrease of yield in their studied populations. It is worth noting that, among 256 RILs of QH34 × JD17, one RIL family QJ163 contains QTL favorable alleles that simultaneously increased soybean 100 seed weight, oil content, and protein plus oil content in Beijing. Moreover, QJ186 and QJ235 contain QTL favorable alleles that could simultaneously increase seed oil, protein, protein plus oil content, and 100 seed weight as well when grown in Sanya. Those families could be used as the excellent germplasms for soybean breeding.
Four candidate genes response for a stable QTL related to 100-seed weight
A QTL identified in all three locations was declared to be stable. In this study, only QTLs related to 100-seed weight on Chr 4 were detected in each three locations, and covers 2.65-Mb interval (3.64–6.29 Mb); there are 296 genes in this region. Coincidentally, the growth adaptability–related J gene, located around 4.1 Mb (4.07–4.08 Mb) on Chr 4, is a member of 296 genes (Lu et al. 2017). Comparing the published genome data of QH34 and JD17 (Liu et al. 2020), no polymorphism was detected in J gene between the two parents, which may explain why the 100-seed weight is not affected by the environmental changes. Pedigree research on the J gene in QH34 showed that this haplotype came from 58 to 161, Feng Di Huang and Ju Xuan No.23 (Table S1).
To identify the candidate genes that related to the soybean 100-seed weight, RNA-seq analysis of the different developed seeds for two parental lines was performed. According to soybean seed establishment and development, differential expressed genes (DEGs) between two parental lines of QH34 and JD17 were extracted, combine with the QTLs that contributable for 100-seed weight, we discovered that only one DEG was identified in stage I and 23 DEGs were detected in stage III, and no DEG detected in stage II. In previous report, the seed formation is a process of cell division to cell number increasing, and cell elongation to cell volume enlargement (Liu et al. 2007). Here, we focused on DEGs that carry sequence variation (SNP variation or insertion-deletion) in the QTL of interested, combine with gene function annotation, four candidate genes of Glyma.04G047800, Glyma.04G051200, Glyma.04G062400, and Glyma.04G073900, involved in the processes of cell development and seed development were identified. The expression pattern of those four genes from stage I to III was totally different, which may explained by involving in different pathways (Fig. 7). Glyma.04G047800 encodes an APETALA2 (AP2)-like factor that has been reported in Arabidopsis, it plays an important role in the regulation of floral organs in Arabidopsis, also regulates the seed development (Jofuku et al. 1994). Glyma.04G051200 is involved in the process of cell differentiation and cell wall modification, and it may contributed to the 100-seed weight by increasing the cell numbers and seed size. Glyma.04G062400 encodes a chromatin remodeling factor CHD3 (PICKLE), a protein that regulates cytokinin signaling in late processes, of which cytokinin has proved to regulate the seed size by regulating the growth of embryonic cells during seed development (Furuta et al. 2011; Werner et al. 2003; Riefler et al. 2006). Glyma.04G073900 is a homolog of AT4G31160, which encodes a DDB1-CUL4 ASSOCIATED FACTOR1 (DCAF1) protein capable of interacting with DDB1 and associating with CUL4, likely as part of a nuclear E3 ubiquitin ligase complex (Zhang et al. 2008). Previous study has proven that proteins related to the ubiquitin pathway are involved in regulating plant seed size (Li et al. 2008). More molecular and biology experiments were required for further evidence of the functions in terms of above four candidates.
Heat map and gene expression pattern of candidates detected in the common 100-seed weight-related QTL detected in three locations. a Heat map of the genes expression detected by RNA-seq analysis (FPKM) during seed development. b, c, d, e Gene expression pattern of Glyma.04G047800, Glyma.04G051200, Glyma.04G062400, and Glyma.04G073900
Implications for breeding
Through pedigree analysis in the condition of the clear genetic background, we can find the origin of QTL favorable alleles, and clearly see the process of the QTL favorable alleles pyramiding into the offspring. In the breeding process, we should achieve the diversity of breeding materials, so that with the pyramiding of excellent alleles, the progenies will become an aggregation of excellent genes.
The QTLs (such as qNP3-1, qSHW4-1, qNHW4-1) identified in previous studies indicate that such excellent alleles widely existed in multiple accessions and they may be retained in the breeding process through phenotypic value selection. We also found QTLs (such as qSP11-1, qNO5-1, qNPplusO11-1) that are undetected in previous studies, indicating that such QTL favorable alleles are unique in the RILs derived from QH34 and JD17 and these types of excellent alleles are relatively rare in the soybean population.
As we all know, there is a bottleneck effect in the domestication process, which causes rare alleles that are more likely to be lost than the widespread alleles, and eventually lead to a smaller gene pool. It is widely known that reduction in genetic diversity may result in the population that is more susceptible to disease. What’s more, in the breeding process, breeders tended to select minority excellent germplasm, which caused the germplasm to be used too frequently to result in the loss of genetic diversity of the varieties in production. The narrow genetic basis of varieties has limited the further improvement of quality and yield, which is essential to maintain the increasing population. Fortunately, the discovery and utilization of these rare alleles can effectively increase population genetic diversity and broaden genetic basis of varieties, so that there would be higher levels of standing genetic variation, making it more likely that some individuals would happen to have a gene variant that conferred the ability to increase quality and yield.
All in all, only by combining the diversity and rarity of germplasm resources can we breed varieties with wide adaptability and high quality.
References
Brummer EC, Graef GL, Orf J, Wilcox JR, Shoemaker RC (1997) Mapping QTL for seed protein and oil content in eight soybean populations. Crop Sci 37:370–378
Chapman A, Pantalone VR, Ustun A, Allen FL, Landau-Ellis D, Trigiano RN, Gresshoff PM (2003) Quantitative trait loci for agronomic and seed quality traits in an F2 and F4:6 soybean population. Euphytica 129:387–393
Diers BW, Keim P, Fehr WR, Shoemaker RC (1992) RFLP analysis of soybean seed protein and oil content. Theor Appl Genet 83:608–612
Furuta K, Kubo M, Sano K, Demura T, Fukuda H, Liu YG, Shibata D, Kakimoto T (2011) The ckh2/pkl chromatin remodeling factor negatively regulates cytokinin responses in Arabidopsis calli. Plant Cell Physiol 52:618–628
Hacisalihoglu G, Burton A, Gustin J, Eker S, Asikli S, Heybet E, Ozturk L, Cakmak I, Yazici A, Burkey K, Orf J, Settles A (2018) Quantitative trait loci associated with soybean seed weight and composition under different phosphorus levels. JIPB 3:232–241
Han Y, Li D, Zhu D, Li H, Li X, Teng W, Li W (2012) QTL analysis of soybean seed weight across multi-genetic backgrounds and environments. Theor Appl Genet 125:671–683
Hyten DL, Pantalone VR, Sams CE, Saxton AM, Landau-Ellis D, Stefaniak TR, Schmidt ME (2004) Seed quality QTL in a prominent soybean population. Theor Appl Genet 109:552–561
Jofuku KD, den Boer BGW, Van Montagu M, Okamuro JK (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6:1211–1225
Lee SH, Bailey MA, Mian MAR, Carter TE Jr, Shipe ER, Ashley DA, Parrott WA, Hussey RS, Boerma HR (1996) RFLP loci associated with soybean seed protein and oil content across populations and locations. Theor Appl Genet 93:649–657
Li H, Ye G, Wang J (2007) A modified algorithm for the improvement of composite interval mapping. Genetics 175:361–374
Li YH, Zheng LY, Corke F, Smith C, Bevan MW (2008) Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev 22:1331–1336
Li YH, Reif JC, Hong HL, Li HH, Liu ZX, Ma YS, Li J, Tian Y, Li YF, Li WB, Qiu LJ (2018) Genome-wide association mapping of QTL underlying seed oil and protein contents of a diverse panel of soybean accessions. Plant Science 266:95–101
Liu X, Jin J, Wang G, Herbert SJ (2007) Regulation of soybean seed size by eco-physiological factors. Soybean Sci 26:607–612 (in Chinese)
Liu Y, Du H, Li P, Shen Y, Peng H, Liu S, Zhou G, Zhang H, Liu Z, Shi M, Huang X, Li Y, Zhang M, Wang Z, Zhu B, Han B, Liang C, Tian Z (2020) Pan-genome of wild and cultivated soybeans. Cell 182:162–176
Lu SJ, Zhao XH, Hu YL, Liu SL, Nan HY, Li XM, Fang C, Cao D, Shi XY, Kong LP, Su T, Zhang FG, Li SC, Wang Z, Yuan XH, Cober ER, Weller JL, Liu BH, Hou XL, Tian ZX, Kong FJ (2017) Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat Genet 49:773–779
Mansur LM, Orf JH, Chase K, Jarvik T, Cregan PB, Lark KG (1996) Genetic mapping of agronomic traits using recombinant inbred lines of soybean. Crop Sci 36:1327–1336
Mao T, Jiang Z, Han Y, Teng W, Zhao X, Li W (2013) Identification of quantitative trait loci underlying seed protein and oil contents of soybean across multi-genetic backgrounds and environments. Plant Breed 132:630–641
Mian MAR, Bailey MA, Tamulonis JP, Shipe ER, Carter TE Jr, Parrott WA, Ashley DA, Hussey RS, Boerma HR (1996) Molecular markers associated with seed weight in two soybean populations. Theor Appl Genet 93:1011–1016
Miao L, Yang S, Zhang K, He J, Wu C, Ren Y, Gai J, Li Y (2020) Natural variation and selection in GmSWEET39 affect soybean seed oil content. New Phytol 225:1651–1666
Orf JH, Chase K, Jarvik T, Mansur LM, Cregan PB, Adler FR, Lark KG (1999) Genetics of soybean agronomic traits: I. comparison of three related recombinant inbred populations. Crop Sci 39:1642–1651
Pathan SM, Vuong T, Clark K, Lee JD, Shannon JG, Roberts CA, Ellersieck MR, Burton JW, Cregan PB, Hyten DL, Nguyen HT, Sleper DA (2013) Genetic mapping and confirmation of quantitative trait loci for seed protein and oil contents and seed weight in soybean. Crop Sci 53:765–774
Rajcan I, Hou G, Weir AD (2005) Advances in breeding of seed-quality traits in soybean. J. Crop Improv 14:145–174
Reinprecht Y, Poysa VW, Yu K, Rajcan I, Ablett GR, Pauls KP (2006) Seed and agronomic QTL in low linolenic acid, lipoxygenase-free soybean [Glycine max (L.) Merrill] germplasm. Genome 49:1510–1527
Riefler M, Novak O, Strnad M, Schmülling T (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18:40–54
Sebolt AM, Shoemaker RC, Diers BW (2000) Analysis of a quantitative trait locus allele from wild soybean that increases seed protein concentration in soybean. Crop Sci 40:1438–1444
Specht JE, Chase K, Macrander M, Graef GL, Chung J, Markwell JP, Germann M, Lark KG, Orf JH (2001) Soybean response to water: a QTL analysis of drought tolerance. Crop Sci 41:493–509
Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15:2532–2550
Wilcox JR (1998) Increasing seed protein in soybean with eight cycles of recurrent selection. Crop Sci 38:1536–1540
Wilcox JR, Zhang GD (1997) Relationships between seed yield and seed protein in determinate and indeterminate soybean populations. Crop Sci 37:361–364
Yan L, Hofmann N, Li S, Ferreira ME, Song B, Jiang G, Ren S, Quigley C, Fickus E, Cregan P, Song Q (2017) Identification of QTL with large effect on seed weight in a selective population of soybean with genome-wide association and fixation index analyses. BMC Genomics 18:529
Yang K, Moon JK, Jeong N, Chun HK, Kang ST, Back K, Jeong SC (2011) Novel major quantitative trait loci regulating the content of isoflavone in soybean seeds. Gen Genom 33:685–692
Yang H, Wang W, He Q, Xiang S, Tian D, Zhao T, Gai J (2019) Identifying a wild allele conferring small seed size, high protein content and low oil content using chromosome segment substitution lines in soybean. Theor Appl Genet 132(10):2793–2807
Zhang Y, Feng SH, Chen FF, Chen HD, Wang J, McCall C, Xiong Y, Deng XW (2008) Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. Plant Cell 20:1437–1455
Zhang D, Zhang H, Hu Z, Chu S, Yu K, Lv L, Yang Y, Zhang X, Chen X, Kan G, Tang Y, An YQC, Yu D (2019) Artificial selection on GmOLEO1 contributes to the increase in seed oil during soybean domestication. PLoS Genet 15(7):e1008267
Acknowledgements
We thank Feng Lin at Michigan State University for suggestions for paper modification.
Funding
This research was financially supported by the National Natural Science Foundation of China (Grant nos. 31871708 and 32072089), the Fundamental Research Funds for the Central Universities (Grant nos.2020TC178), and the Recruitment Program of Global Experts.
Author information
Authors and Affiliations
Contributions
L. Sun conceived and designed the research; L. Li, J. Hou, J. An, and Q. Han performed the experiments; W. Huang, Q. Hu and X. Li analyzed the data; J. An, Y. Zhang and Y. Wu constructed the RIL population; W. Huang wrote the manuscript with input from L. Sun, L. Li, D. Zhang, R. Xu and J. Wang. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval
The experiments were performed in compliance with the current laws of China.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Table S1
(XLSX 10 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Huang, W., Hou, J., Hu, Q. et al. Pedigree-based genetic dissection of quantitative loci for seed quality and yield characters in improved soybean. Mol Breeding 41, 14 (2021). https://doi.org/10.1007/s11032-021-01211-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-021-01211-6