Abstract
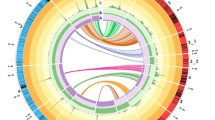
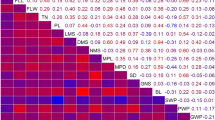
Genetic mapping in ornamental plant species has lagged behind crop plants from other sectors of agriculture. Here, we describe the generation of a genetic linkage map for the important herbaceous ornamental crop petunia and the identification of QTL for several key crop timing and quality traits, including plant development rate, days to flower and flower number. An F2 population derived from a cross between the progenitor species of cultivated petunia, P. integrifolia × P. axillaris, exhibited transgressive segregation for a broad panel of crop timing and quality traits. A genetic linkage map comprised of 75 simple sequence repeat and six cleaved amplified polymorphic sequence markers spanning 359.1 cM across seven linkage groups was developed and utilized to identify 24 QTL for ten crop timing and quality traits. These included QTL explaining 26.3, 25.9, 26.2 and 43 % of the observed phenotypic variation for flower length, branch number, internode length and the number of flower buds on the primary shoot, respectively. These data provide a foundation for understanding the genetic control of critical traits and identify molecular markers with potential utility to facilitate gene discovery in petunia.


Similar content being viewed by others
References
Abe H, Nakano M, Nakatsuka A, Nakayama M, Koshioka M, Yamagashi M (2002) Genetic analysis of floral anthocyanin pigmentation traits in Asiatic hybrid lily using molecular linkage maps. Theor Appl Genet 105:1175–1182
Ando T, Hashimoto G (1993) Two new species of Petunia (Solanaceae) from southern Brazil. Bot J Linn Soc 111:265–280
Bartok JW (2001) Energy conservation for commercial greenhouses, 2001 rev. Natural Resource, Agriculture, and Engineering Service, Ithaca
Blanchard MG, Runkle ES (2011) The influence of day and night temperature fluctuations on growth and flowering of annual bedding plants and greenhouse heating cost predictions. J Am Soc Hortic Sci 46:599–603
Bossolini E, Klahre U, Brandenburg A, Reinhardt D, Kuhlemeier C (2011) High resolution linkage maps of the model organism Petunia reveal substantial synteny decay with the related genome of tomato. Genome 54:327–340
Broman KW, Wu H, Sen S, Churchill GA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19:889–890
Cornu A, Farcy E, Mousset C (1989) A genetic basis for variations in meiotic recombination in Petunia hybrida. Genome 32:46–53
Debener T, Mattiesch L (1999) Construction of a genetic linkage map for roses using RAPD and AFLP markers. Theor Appl Genet 99:891–899
Dugo ML, Satovic Z, Millán T, Cubero JI, Rubiales D, Cabrera A, Torres AM (2005) Genetic mapping of QTLs controlling horticultural traits in diploid roses. Theor Appl Genet 111:511–520
Galliot C, Hoballah M, Kuhlemeier C, Stuurman J (2006) Genetics of flower size and nectar volume in Petunia pollination syndromes. Planta 225:203–212
Gerats T, Keukeleire P, Deblaere R, Montagu M, Zethof J (1995) Amplified fragment length polymorphism (AFLP) mapping in Petunia; a fast and reliable method for obtaining a genetic map. Acta Hortic 420:58–61
Griesbach RJ (2007) Petunia. In: Anderson NO (ed) Flower breeding and genetics. Springer, Dordrecht, pp 301–336
Hammer K (1984) Das Domestikationssyndrom. Die Kulturpflanze 32:11–34
Helliwell CA, Chin-Atkins AN, Wilson IW, Chapple R, Dennis ES, Chaudury A (2001) The arabidopsis AMP1 gene encodes a putative glutamate carboxypeptidase. Plant Cell 13:2115–2125
Kawakatsu T, Itoh J-I, Miyoshi K, Kurata N, Alvarez N, Veit B, Nagato Y (2006) PLASTOCHRON2 regulates leaf initiation and maturation in rice. Plant Cell 18:612–625
Klahre U, Gurba A, Hermann K, Saxenhofer M, Bossolini E, Guerin PAM, Kuhlemeier C (2011) Pollinator choice in petunia depends on two major genetic loci for floral scent production. Curr Biol 21:730–739
Kosambi DD (1944) The estimation of map distance from recombination values. Ann Eugen 12:172–175
Lande R, Thompson R (1990) Efficiency of marker-assisted selection in the improvement of quantitative traits. Genetics 124:743–756
Mather K, Edwardes PMJ (1943) Specific differences in Petunia: III. Flower colour and genetic isolation. J Genet 45:243–260
Meyer RS, Purugganan MD (2013) Evolution of crop species: genetics of domestication and diversification. Nat Rev Genet 14:840–852
Miyoshi K, Ahn B-O, Kawakatsu T, Ito Y, Itoh J-I, Nagato Y, Kurata N (2004) PLASTOCHRON1, a timekeeper of leaf initiation in rice, encodes cytochrome P450. PNAS 101:875–880
Mohan M, Nair S, Bhagwat A, Krishna TG, Yano M, Bhatia CR, Sasaki T (1997) Genome mapping, molecular markers and marker-assisted selection in crop plants. Mol Breed 3:87–103
Prigge MJ, Wagner DR (2001) The arabidopsis SERRATE gene encodes a zinc-finger protein required for normal shoot development. Plant Cell 13:1263–1280
Reed JW, Nagpal P, Poole DS, Furuya M, Chory J (1993) Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5:147–157
Robert N, Farcy E, Cornu A (1991) Genetic control of meiotic recombination in Petunia hybrida: dosage effect of gene Rm1 on segments Hf1-Lg1 and An2-Rt; role of modifiers. Genome 34:515–523
Shahin A, Arens P, van Heusden AW, van der Linden G, van Kaauwen M, Schouten HJ, van de Weg WE, Visser RGF, van Tuyl JM (2011) Genetic mapping in Lilium: mapping of major genes and quantitative trait loci for several ornamental traits and disease resistances. Plant Breed 130:372–382
Sims TL, Gomez AB, Delledonne M, Gerats T, Mitrick J, Mueller L, Pezzotti M, Quattrocchio F, Yang B (2012). Sequencing and comparison of the genomes of Petunia inflata and Petunia axillaris. In: Plant and Animal Genome XX Conference (abstract) https://pag.confex.com/pag/xx/webprogram/Paper1786.html. Accessed 26 Oct 2014
Stehmann JR, Lorenz-Lemke AP, Freitas LB, Semir J (2009) The genus Petunia. In: Gerats T, Strommer J (eds) Petunia evolutionary, developmental and physiological genetics, Springer, New York, pp 1–28. doi:10.1007/978-0-387-84796-2
Strommer J, Gerats AGM, Sanago M, Molnar SJ (2000) A gene-based RFLP map of petunia. Theor Appl Genet 100:899–905
Strommer J, Peters J, Zethof J, de Keukeleire P, Gerats T (2002) AFLP maps of Petunia hybrida: building maps when markers cluster. Theor Appl Genet 105:1000–1009
Strommer J, Peters JL, Gerats T (2009) Genetic recombination and mapping in Petunia. In: Gerats T, Strommer J (eds) Petunia. Springer, New York, pp 325–341
Stuurman J, Hoballah ME, Broger L, Moore J, Basten C, Kuhlemeier C (2004) Dissection of floral pollination syndromes in petunia. Genetics 168:1585–1599
Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
Ten Hoopen R, Robbins TP, Fransz PF, Montijn BM, Oud O, Gerats AGM, Nanninga N (1996) Localization of T-DNA insertions in petunia by fluorescence in situ hybridization: physical evidence for suppression of recombination. Plant Cell 8:823–830
Tychonievich J, Wangchu L, Barry C, Warner RM (2013) Utilizing wild species for marker-assisted selection of crop timing and quality traits in Petunia. Acta Hortic 1000:465–469
Van Ooijen JW (2006) JoinMap® 4, Software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen, Netherlands
Veit B, Briggs SP, Schmidt RJ, Yanofsky MF, Hake S (1998) Regulation of leaf initiation by the terminal ear 1 gene of maize. Nature 393:166–168
Vlaming P, Gerats AGM, Wiering H, Wijsman HJW, Cornu A, Farcy E, Maizonnier D (1984) Petunia hybrida: a short description of the action of 91 genes, their origin and their map location. Plant Mol Biol Rep 2:21–42
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wang Y, Wang X, McCubbin AG, Kao T (2003) Genetic mapping and molecular characterization of the self-incompatibility (S) locus in Petunia inflata. Plant Mol Biol 53:565–580
Wang S, Basten CJ, Zeng Z-B (2012) Windows QTL cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh. http://statgen.ncsu.edu/qtlcart/WQTLCart.htm. Accessed 22 Oct 2014
Warner RM, Walworth AE (2010) Quantitative inheritance of crop timing traits in interspecific hybrid Petunia populations and interactions with crop quality parameters. J Hered 101:308–316
Watanabe H, Ando T, Iida S, Suzuki A, Buto K-I, Tsukamoto T, Hashimoto G, Marchesi E (1996) Cross compatibility of Petunia cultivars and P. axillaris with native taxa of Petunia in relation to their chromosome number. J Jpn Soc Hortic Sci 65:625–634
Watanabe H, Ando T, Tsukamoto T, Hashimoto G, Marchesi E (2001) Cross-compatibility of Petunia exserta with other Petunia taxa. J Jpn Soc Hortic Sci 70:33–40
Zubko E, Adams CJ, Macháèková I, Malbeck J, Scollan C, Meyer P (2002) Activation tagging identifies a gene from Petunia hybrida responsible for the production of active cytokinins in plants. Plant J 29:797–808
Acknowledgments
We thank Mike Olrich for technical assistance. This research was supported by the US Dept. of Agriculture (USDA) Specialty Crop Block Grant Program and the USDA Specialty Crop Research Initiative (award 2011-01508). R. M .W. and C. S. B. are supported in part by Michigan AgBioResearch and through USDA National Institute of Food and Agriculture, Hatch project numbers MICL02121 and MICL02265.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Vallejo, V.A., Tychonievich, J., Lin, WK. et al. Identification of QTL for crop timing and quality traits in an interspecific Petunia population. Mol Breeding 35, 2 (2015). https://doi.org/10.1007/s11032-015-0218-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-015-0218-4