Abstract
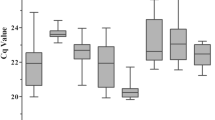
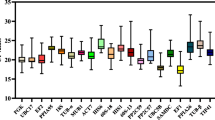
Quantitative real-time RT-PCR (RT-qPCR) is a technology that can be used to analyze the abundance of gene expression. Reference genes, which are assumed to remain at constant levels in different tissues at various developmental stages and photoperiodic treatments, were selected to analyze the expression levels of flowering time genes and floral development genes. Using digital gene expression technology, nine reference genes with moderate expression in the leaves of Chrysanthemum lavandulifolium at the juvenile phase (CK1) and the squaring stage (W1) were selected as the candidate reference genes for further study. A total of 115 biological samples of C. lavandulifolium were analyzed, including different tissues under various developmental stages and leaves with varied photoperiodic treatments. The stability of the nine reference genes was slightly variable across the samples, but MTP, SKIP16 and PGK were the most stable genes overall. In addition, the relative expression level of ClFT in different tissues of plants with the competence to flower was analyzed to verify the reference genes selected in this study. These studies provide a guide for selecting reference genes for analyzing the expression pattern of flowering time genes and floral development genes in C. lavandulifolium.


Similar content being viewed by others
Abbreviations
- ACT :
-
Actin
- CTAB:
-
Cethyltrimethylammonium bromide
- Cq:
-
Quantification cycle
- DD:
-
Constant darkness
- EF1b :
-
Elongation factor 1-beta
- FT :
-
Flowering Locus T
- LD:
-
Long-day
- LL:
-
Constant light
- MTP :
-
Metalloprotease
- NF:
-
Normalization factor
- PGK :
-
Phosphoglycerate kinase
- psaA :
-
Photosynthesis-related plastid gene representing photosystem I
- RPKM:
-
Reads Per Kb per Million reads
- RT-qPCR:
-
Quantitative real-time RT-PCR
- SAND :
-
SAND family protein
- SD:
-
Short-day
- SKIP16 :
-
SKP1/ASK-interacting protein 16
- TUA :
-
α-Tubulin
- TIP41 :
-
TIP41-like family protein
- T m :
-
Melting temperature
References
Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64(15):5245–5250
Anderson LE, Carol AA (2005) Enzyme co-localization in the pea leaf cytosol: 3-P-glycerate kinase, glyceraldehyde-3-P dehydrogenase, triose-P isomerase and aldolase. Plant Sci 169(3):620–628
Artico S, Nardeli SM, Brilhante O, Grossi-de-Sa MF, Alves-Ferreira M (2010) Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biol 10(1):49–60
Brunner AM, Yakovlev IA, Strauss SH (2004) Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol 4(1):14–20
Bustin SA (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25(2):169–193
Chen FH (1957) Origin of Chinese chrysanthemum. Chry Dahlia 7:264–265
Chen JY (1984) Studies on the origin of Chinese florist’s chrysanthemum. Acta Hortic 167:349–362
Coker JS, Davies E (2003) Selection of candidate housekeeping controls in tomato plants using EST data. Biotechniques 35(4):740–749
Cruz F, Kalaoun S, Nobile P, Colombo C, Almeida J, Barros LMG, Romano E, Grossi-de-Sá MF, Vaslin M, Alves-Ferreira M (2009) Evaluation of coffee reference genes for relative expression studies by quantitative real-time RT-PCR. Mol Breed 23(4):607–616
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139(1):5–17
Dai SL, Wang WK, Huang JP (2002) Advances of researches on phylogeny of Dendranthema and origin of chrysanthemum. J Beijing Forest Univ 24(5/6):230–234
Dai SL, Wang WK, Li MX, Xu YX (2005) Phylogenetic relationship of Dendranthema (DC.) Des Moul. revealed by fluorescent in situ hybridization. J Integr Plant Biol 47(7):783–791
Dheda K, Huggett JF, Chang JS, Kim LU, Bustin SA, Johnson MA, Rook GAW, Zumla A (2005) The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal Biochem 344(1):141–143
Expósito-Rodríguez M, Borges AA, Borges-Pérez A, Pérez JA (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8(1):131–142
Fernandez P, Di Rienzo JA, Moschen S, Dosio GAA, Aguirrezábal LAN, Hopp HE, Paniego N, Heinz RA (2011) Comparison of predictive methods and biological validation for qPCR reference genes in sunflower leaf senescence transcript analysis. Plant Cell Rep 30(1):63–74
Gu CS, Chen SM, Liu ZL, Shan H, Luo HL, Guan ZY, Chen FD (2011) Reference gene selection for quantitative real-time PCR in chrysanthemum subjected to biotic and abiotic stress. Mol Biotechnol 49(2):192–197
Hong SY, Seo PJ, Yang MS, Xiang FN, Park CM (2008) Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol 8(1):112–122
Hu RB, Fan CM, Li HY, Zhang QZ, Fu YF (2009) Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol Biol 10(1):93–104
Huang H, Cao HW, Niu YJ, Dai SL (2012a) Expression analysis of Nudix Hydrolase genes in Chrysanthemum lavandulifolium. Plant Mol Biol Rep 30(4):973–982
Huang H, Niu YJ, Cao HW, Tang XJ, Xia XL, Yin WL, Dai SL (2012b) cDNA-AFLP analysis of salt-inducible genes expression in Chrysanthemum lavandulifolium under salt treatment. J Plant Physiol 169(4):410–420
Huang H, Niu YJ, Yang K, Dai SL (2012c) Isolation and expression analysis of a new reference gene: ClTUA of Chrysanthemum lavandulifolium. J Beijing Forest Univ 34(2):112–117
Huggett J, Dheda K, Bustin S, Zumla A (2005) Real-time RT-PCR normalisation; strategies and considerations. Genes Immun 6(4):279–284
Jain M (2009) Genome-wide identification of novel internal control genes for normalization of gene expression during various stages of development in rice. Plant Sci 176(5):702–706
Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345(2):646–651
Jian B, Liu B, Bi YR, Hou WS, Wu CX, Han TF (2008) Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol Biol 9(1):59–72
Kim BR, Nam HY, Kim SU, Kim SI, Chang YJ (2003) Normalization of reverse transcription quantitative-PCR with housekeeping genes in rice. Biotechnol Lett 25(21):1869–1872
Kuijk E, du Puy L, van Tol HT, Haagsman HP, Colenbrander B, Roelen BA (2007) Validation of reference genes for quantitative RT-PCR studies in porcine oocytes and preimplantation embryos. BMC Dev Biol 7(1):58–65
Lee JM, Roche JR, Donaghy DJ, Thrush A, Sathish P (2010) Validation of reference genes for quantitative RT-PCR studies of gene expression in perennial ryegrass (Lolium perenne L.). BMC Mol Biol 11(1):8–21
Li HP, Qin YX, Xiao XH, Tang CR (2011) Screening of valid reference genes for real-time RT-PCR data normalization in Hevea brasiliensis and expression validation of a sucrose transporter gene HbSUT3. Plant Sci 181(2):132–139
Lin YL, Lai ZX (2010) Reference gene selection for qPCR analysis during somatic embryogenesis in longan tree. Plant Sci 178(4):359–365
Løvdal T, Lillo C (2009) Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Anal Biochem 387(2):238–242
Ma YP, Fang XH, Chen F, Dai SL (2008) DFL, a FLORICAULA/LEAFY homologue gene from Dendranthema lavandulifolium is expressed both in the vegetative and reproductive tissues. Plant Cell Rep 27(4):647–654
Mallona I, Lischewski S, Weiss J, Hause B, Egea-Cortines M (2010) Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biol 10(1):4–14
Maroufi A, Van Bockstaele E, De Loose M (2010) Validation of reference genes for gene expression analysis in chicory (Cichorium intybus) using quantitative real-time PCR. BMC Mol Biol 11(1):15–26
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5(7):621–628
Oda A, Narumi T, Li T, Kando T, Higuchi Y, Sumitomo K, Fukai S, Hisamatsu T (2012) CsFTL3, a chrysanthemum FLOWERING LOCUS T-like gene, is a key regulator of photoperiodic flowering in chrysanthemums. J Exp Bot 63(3):1461–1477
Paolacci AR, Tanzarella OA, Porceddu E, Ciaffi M (2009) Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol Biol 10(1):11–37
Reid KE, Olsson N, Schlosser J, Peng F, Lund ST (2006) An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol 6(1):27–37
Remans T, Smeets K, Opdenakker K, Mathijsen D, Vangronsveld J, Cuypers A (2008) Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 227(6):1343–1349
Rozen S, Skaletsky H (1999) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols in the series methods in molecular biology. Humana Press, Totowa, pp 365–386
Schmidt GW, Delaney SK (2010) Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol Genet Genomics 283(3):233–241
Silberberg G, Baruch K, Navon R (2009) Detection of stable reference genes for real-time PCR analysis in schizophrenia and bipolar disorder. Anal Biochem 391(2):91–97
Suzuki Y, Mae T, Makino A (2008) RNA extraction from various recalcitrant plant tissues with a cethyltrimethylammonium bromide-containing buffer followed by a acid guanidium thiocyanate-phenol-chloroform treatment. Biosci Biotechnol Biochem 72(7):1951–1953
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3(7):research0034.0031–0034.0011
Wang LL, Fu JX, Dai SL (2011) ClFT, A Novel Homologues Gene of FLOWERING LOCUS T from Chrysanthemum lavandulifolium. Mol Plant Breed 9:1062–1064
Zhang LJ, Dai SL (2009) Research Advance on Germplasm Resources of Chrysanthemum × morifolium. Chin Bull Bot 44(5):526–535
Acknowledgments
This work was supported by the National Natural Science Foundation of China (30871726 and 31071823), the Forestry Administration of China National Public Project (200904050) and the Chinese 863 Project (2007AA021403).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fu, J., Wang, Y., Huang, H. et al. Reference gene selection for RT-qPCR analysis of Chrysanthemum lavandulifolium during its flowering stages. Mol Breeding 31, 205–215 (2013). https://doi.org/10.1007/s11032-012-9784-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-012-9784-x