Abstract
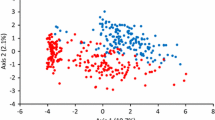
Drought often delays developmental events so that plant height and above-ground biomass are reduced, resulting in yield loss due to inadequate photosynthate. In this study, plant height and biomass measured by the Normalized Difference Vegetation Index (NDVI) were used as criteria for drought tolerance. A total of 305 lines representing temperate, tropical and subtropical maize germplasm were genotyped using two single nucleotide polymorphism (SNP) chips each containing 1536 markers, from which 2052 informative SNPs and 386 haplotypes each constructed with two or more SNPs were used for linkage disequilibrium (LD) or association mapping. Single SNP- and haplotype-based LD mapping identified two significant SNPs and three haplotype loci [a total of four quantitative trait loci (QTL)] for plant height under well-watered and water-stressed conditions. For biomass, 32 SNPs and 12 haplotype loci (30 QTL) were identified using NDVIs measured at seven stages under the two water regimes. Some significant SNP and haplotype loci for NDVI were shared by different stages. Comparing significant loci identified by single SNP- and haplotype-based LD mapping, we found that six out of the 14 chromosomal regions defined by haplotype loci each included at least one significant SNP for the same trait. Significant SNP haplotype loci explained much higher phenotypic variation than individual SNPs. Moreover, we found that two significant SNPs (two QTL) and one haplotype locus were shared by plant height and NDVI. The results indicate the power of comparative LD mapping using single SNPs and SNP haplotypes with QTL shared by plant height and biomass as secondary traits for drought tolerance in maize.

Similar content being viewed by others
Abbreviations
- abph1:
-
Aberrant phyllotaxy1
- ALDH:
-
Aldehyde dehydrogenase
- ASI:
-
Anthesis–silking interval
- CAAS:
-
Chinese Academy of Agricultural Sciences
- CIMMYT:
-
International Maize and Wheat Improvement Center
- DAP:
-
Days after planting
- DT chip:
-
Drought tolerance candidate gene based chip
- GWAS:
-
Genome-wide association study
- HP:
-
Haplotype
- IL:
-
Introgression line
- LD:
-
Linkage disequilibrium
- MAF:
-
Minor allele frequency
- MLM:
-
Mixed linear model
- NDVI:
-
Normalized Difference Vegetation Index
- NIR:
-
Near infrared region
- OPA:
-
Oligo pool assay
- PC:
-
Principal component
- PCA:
-
Principal component analysis
- PH:
-
Plant height
- PIC:
-
Polymorphism information content
- RA chip:
-
Random candidate gene based chip
- SNP:
-
Single nucleotide polymorphism
- VIS:
-
Visible region
- WS:
-
Water-stressed
- WW:
-
Well-watered
References
Aranzana MJ, Kim S, Zhao K, Bakker E, Horton M, Jakob K, Lister C, Molitor J, Shindo C, Tang C, Toomajian C, Traw B, Zheng H, Bergelson J, Dean C, Marjoram P, Nordborg M (2005) Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genet 1:e60
Bänziger M, Araus J (2007) Recent advances in breeding maize for drought and salinity stress tolerance. In: Jenks MA, Hasegawa PM, Jain SM (eds) Advances in molecular breeding toward drought and salt tolerant crops. Springer, Netherlands, pp 587–601
Berke T, Rocheford T (1995) Quantitative trait loci for flowering, plant and ear height, and kernel traits in maize. Crop Sci 35:1542–1549
Bolanos J, Edmeades GO, Martinez L (1993) Eight cycles of selection for drought tolerance in tropical maize. III. Responses in drought-adaptive physiological and morphological traits. Field Crops Res 31:269–286
Bruce WB, Edmeades GA, Barker TC (2002) Molecular and physiological approaches to maize improvement for drought tolerance. J Exp Bot 53:13–25
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Casa AM, Pressoira G, Brown PJ, Mitchell SE, Rooney WL, Tuinstrac MR, Franks CD, Kresovich S (2008) Community resources and strategies for association mapping in sorghum. Crop Sci 48:30–40
Casati P, Stapleton AE, Blum JE, Walbot V (2006) Genome-wide analysis of high-altitude maize and gene knockdown stocks implicates chromatin remodeling proteins in response to UV-B. Plant J 46:613–627
CIMMYT (1994) QTL data for populations Ki3 x CML139 and CML131 x CML67. http://www.agron.missouri.edu/locus.html
CIMMYT Applied Molecular Genetics Laboratory (2003) Laboratory protocols, 3rd edn. Mexico, DF, pp 7–11
Edmeades GO, Bolaños J, Chapman SC (1997) Value of secondary traits in selecting for drought tolerance in tropical maize. In: Edmeades GO, Bänziger M, Mickelson HR, Peña-Valdivia CB (eds) Developing drought and low-N tolerant maize. CIMMYT, Mexico, pp 222–234
Edmeades GO, Bolaños J, Elings A, Ribaut JM, Bänziger M, Westgate ME (2000) The role and regulation of the anthesis-silking interval in maize. In: Westgate M, Boote K (eds) Physiology and modeling kernel set in maize. Proceedings of ASA-CSSA-SSSA Meeting, Baltimore, USA. 17–22 October 1998, CSSA Special Publication Number 29, 43–75
Edmeades GO, Bolaños J, Hernandez M, Bello S (1993) Causes for silk delay in a lowland tropical maize population. Crop Sci 33:1029–1035
Fan JB, Gunderson KL, Bibikova M, Yeakley JM, Chen J, Garcia EW, Lebruska LL, Laurent M, Shen R, Barker D (2006) Illumina universal bead arrays. Methods Enzymol 410:57–73
Hamblin MT, Warburton ML, Buckler ES (2007) Empirical comparison of simple sequence repeats and single nucleotide polymorphisms in assessment of maize diversity and relatedness. PLoS ONE 12:e1367
Hao Z, Liu X, Li X, Xie C, Li M, Zhang D, Zhang S, Xu Y (2009) Identification of quantitative trait loci for drought tolerance at seedling stage by screening a large number of introgression lines in maize. Plant Breed 128:337–341
Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y, Li C, Zhu C, Lu T, Zhang Z, Li M, Fan D, Guo Y, Wang A, Wang L, Deng L, Li W, Lu Y, Weng Q, Liu K, Huang T, Zhou T, Jing Y, Li W, Lin Z (2010) Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet 42:961–967
Koester RP, Sisco PH, Stuber CW (1993) Identification of quantitative trait loci controlling days to flowering and plant height in two near isogenic lines of maize. Crop Sci 33:1209–1216
Lafitte R, Blum A, Atlin G (2003) Using secondary traits to help identify drought-tolerant genotypes. In: Fischer KS, Lafitte R, Fukai S, Atlin G, Hardy B (eds) Breeding rice for drought-prone environments. IRRI, Manila
Laval G, San Cristobal M, Chevalet C (2002) Measuring genetic distances between breeds: use of some distances in various short term evolution models. Genet Sel Evol 34:481–507
Lee BH, Johnston R, Yang Y, Gallavotti A, Kojima M, Travencolo BAN, Costa LF, Sakakibara H, Jackson D (2009) Studies of aberrant phyllotaxy1 mutants of maize indicate complex interactions between auxin and cytokinin signaling in the shoot apical meristem. Plant Physiol 150:205–216
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Lu Y, Yan J, Guimarães CT, Taba S, Hao Z, Gao S, Chen S, Li J, Zhang S, Vivek BS, Mugo S, Makumbi D, Shah T, Rong T, Crouch JH, Xu Y (2009) Molecular characterization of global maize breeding germplasm based on genome-wide single nucleotide polymorphisms. Theor Appl Genet 120:93–115
Lu Y, Zhang SH, Shah T, Xie C, Hao Z, Li X, Farkhari M, Ribaut JM, Cao M, Rong T, Xu Y (2010) Joint linkage–linkage disequilibrium mapping is a powerful approach to detecting quantitative trait loci underlying drought tolerance in maize. Proc Natl Acad Sci USA 107:19585–19590
Lu Y, Hao Z, Xie C, Crossa J, Araus JL, Gao S, Vivek BS, Magorokosho C, Mugo S, Makumbi D, Taba S, Pan G, Li X, Rong T, Zhang S, Xu Y (2011a) Comparison of multiple selection criteria for drought resistance through large-scale screening under water-stressed and well-watered environments in maize. Field Crops Res 124:37–45
Lu Y, Shah T, Hao Z, Taba S, Zhang S, Gao S, Liu J, Cao M, Wang J, Prakash AB, Rong T, Xu Y (2011b) Comparative SNP and haplotype analysis reveals a higher genetic diversity and rapider LD decay in tropical than temperate germplasm in maize. PLoS ONE 6(9):e24861
Malosetti M, van der Linden CG, Vosman B, van Eeuwijk FA (2007) A mixed-model approach to association mapping using pedigree information with an illustration of resistance to Phytophthora infestans in potato. Genetics 175:879–889
Messmer R, Fracheboud Y, Bänziger M, Vargas M, Stamp P, Ribaut JM (2009) Drought stress and tropical maize: QTL-by-environment interactions and stability of QTLs across environments for yield components and secondary traits. Theor Appl Genet 119:913–930
Paulsen IT, Pao SS, Saier MH Jr (1998) Major facilitator superfamily. Microbiol Mol Biol Rev 62:1–34
Ribaut JM, Betran J, Monneveux P, Setter T (2009) Drought tolerance in maize. In: Bennetzen JL, Hake SC (eds) Handbook of maize: its biology. Springer, New York, pp 324–325
Setter TL, Yan J, Warburton M, Ribaut JM, Xu Y, Sawkins M, Buckler ES, Zhang Z, Gore MA (2010) Genetic association mapping identifies single nucleotide polymorphisms in genes that affect abscisic acid levels in maize floral tissues during drought. J Exp Bot 62:701–716
Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielsen D, Buckler ES (2001) Dwarf8 polymorphisms associate with variation in flowering time. Nat Genet 28:286–289
Tian F, Bradbury PJ, Brown PJ, Hung H, Sun Q, Flint-Garcia S, Rocheford TR, McMullen MD, Holland JB, Buckler ES (2011) Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat Genet 43:159–162
Tuberosa R, Salvi S (2006) Genomics-based approaches to improve drought tolerance of crops. Trend Plant Sci 11:405–412
Veldboom LR, Lee M (1996) Genetic mapping of quantitative trait loci in maize in stress and nonstress environments: II. Plant height and flowering. Crop Sci 36:1320–1327
Wei Y, Lin M, Oliver DJ, Schnable PS (2009) The roles of aldehyde dehydrogenases (ALDHs) in the PDH bypass of Arabidopsis. BMC Biochem 10:7
Wen W, Mei H, Feng F, Yu S, Huang Z, Wu J, Chen L, Xu X, Luo L (2009) Population structure and association mapping on chromosome 7 using a diverse panel of Chinese germplasm of rice (Oryza sativa L.). Theor Appl Genet 119:459–470
Wolfe DW, Henderson DW, Hsiao TC, Alvino A (1988) Interactive water and nitrogen effects on senescence of maize. I. Leaf area duration nitrogen distribution and yield. Agron J 80:859–864
Yan J, Tang H, Huang YQ, Shi YG, Zheng YL, Li JS (2003) Genomic analysis of plant height in maize through molecular marker. Agric Sci Chin 2(10):1069–1075
Yan J, Shah T, Warburton ML, Buckler ES, McMullen MD, Crouch J (2009a) Genetic characterization and linkage disequilibrium estimation of a global maize collection using SNP markers. PLoS ONE 4:e8451
Yan WG, Li Y, Agrama HA, Luo D, Gao F, Lu X, Ren G (2009b) Association mapping of stigma and spikelet characteristics in rice (Oryza sativa L.). Mol Breed 24:277–292
Yu J, Pressoir G, Briggs WH, Vroh BI, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen MD, Holland JB, Kresovich S, Buckler ES (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38:203–208
Zhu C, Gore M, Buckler ES, Yu J (2008) Status and prospects of association mapping in plants. Plant Genome 1:5–20
Acknowledgments
The SNP chips used in this study were developed through a collaborative project between Cornell University (Ed Buckler and Tim Setter) and CIMMYT (Marilyn Warburton and Jianbing Yan). Maize drought tolerance work was funded by the Rockefeller Foundation, Bill and Melinda Gates Foundation, Generation Challenge Programme, European Community, National High Technology Research and Development Program of China (No. 2009AA10AA03-2) and National Basic Research Program of China (No.2007CB108900).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
11032_2011_9631_MOESM1_ESM.xls
List of 305 maize inbred lines used in the study. Information provided in this supplemental table includes ID, brief name, sample name, origin, adaptation, kernel color, and kernel texture (XLS 155 kb)
11032_2011_9631_MOESM2_ESM.xls
Putative candidate genes for significant loci identified by single SNP-based and haplotype-based linkage disequilibrium (LD) mapping. WW: well-watered; WS: water-stressed. *Significant loci identified by both single SNP- and haplotype-based LD mapping. Gene identification numbers, positions and functions were based on maize B73 genome (http://www.maizesequence.org) and NCBI website (http://www.ncbi.nlm.nih.gov/) (XLS 32 kb)
Rights and permissions
About this article
Cite this article
Lu, Y., Xu, J., Yuan, Z. et al. Comparative LD mapping using single SNPs and haplotypes identifies QTL for plant height and biomass as secondary traits of drought tolerance in maize. Mol Breeding 30, 407–418 (2012). https://doi.org/10.1007/s11032-011-9631-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-011-9631-5