Abstract
Japanese encephalitis is a major threat in developing countries, even the availability of several conventional vaccines, which demand development of more effective vaccines. The present study used propred I and Immune Epitope Database Artificial Neural Network (ANN) algorithm (IEDB-ANN) to identify the conserve and promiscuous T cell epitopes from JEV proteome followed by structure based analysis of potential epitopes. Among all identified 102 epitopes, ten epitope were promiscuous but two epitopes of glycoprotein viz. 55LVTVNPFVA63 and 38IPIVSVASL46 were found most promiscuous, highly conserved and high population coverage in comparison of known antigenic positive control peptides. The B cell epitopes of glycoprotein also share these two T cell epitopes revealed by BCPred algorithm which can be a basis to confer the protection by neutralizing antibody combined with an effective cell-mediated response. Further, Autodock 4.2 and NAMD–VMD molecular dynamics simulation were used for docking and molecular dynamics simulation respectively, to validate epitope and allele complex binding stability. The 3D structure models were generated for epitopes and corresponding HLA allele by Pepstr and Modeller 9.10 respectively. Epitope LVTVNPFVA–B5101 allele complex showed best energy minimization and stability over the time window during simulation. Here we also present the binding sequel of epitope LVTVNPFVA and its eventual transport through cTAP1 (PDB ID: 1JJ7) revealed by Autodock 4.2, which is an essential path for HLA class I binding epitopes to elicit immune response. The docking experiment of epitope LVTVNPFVA and cTAP1 very well show a 2 H-bond with a binding energy of −1.88 kcal/mol and other binding state of epitope forming no H-bond with a binding energy of −1.13 kcal/mol in the lower area of cTAP1 cavity. These results show a smooth pass through of the epitope across the channel of cTAP1. Overall, identified peptides have potential application in the design and development of short peptide based vaccines and diagnostic agents for Japanese encephalitis.
Similar content being viewed by others
Introduction
Japanese encephalitis virus (JEV) is a major human pathogenic flavivirus, which is endemic in mostly developing counties. The JEV infections causes severe brain encephalitis in human being with the help of vetor Culex tritaeniorhynchus and Culex. visnui mosquitoes. As reported 67,900 clinical cases of JE occur annually, with approximately 13,600–20,400 deaths, in spite of widespread availability of vaccine (WHO 2015). JEV is single stranded RNA virus of 11,000 nucleotides which translated into three structural proteins and seven non structural proteins (Schiøler et al. 2007). Vaccination only the key to prevent the JEV infection because therapeutic treatment of JE not available yet and vetor (mosquito) control in developing countries is unsuccessfull (Hoke et al. 1992; Igarashi 2002). Therefore the vaccination only become a major thrust option to win over the JEV infections.
Currently, a number of vaccines have been developed for JE, but among them only inactivated mouse brain derived Nakayama strain vaccine is the most accepted vaccine (Barrett 1997; Monath 2002). Recently, Vero cell derived JE vaccine (IXIARO) is giving more promising immune response against JEV in comparison of inactivated vaccine (Erra et al. 2012, 2013) With concern of developing countries, these vaccines have various problems viz. not economic, production shortage and adverse effects (Shlim and Solomon 2002; Okabe 2005), all this leads to the serious requirement to develop more compatible and economically effective vaccines. Immunoinformatics techniques opened a new avenue to screen novel epitopes for development of much better and cost effective vaccines than conventional vaccines. Conventional killed or live attenuated vaccine approaches are not viable for pathogens which are antigenically diverse and not cultivable in laboratory (Singh and Mishra 2016). The option of immunoinformatics approach to develop epitope based vaccine would be rather more effective than conventional vaccines which provides more specific immunity, less time consuming and is devoid of side effects in contrast of entire viral proteins vaccines (De Groot et al. 2002; Sharma and Kumar 2010). In addition to these potentials, the major benefit of epitope based vaccine is the ability to deliver high doses of potential immunogen at lower cost (Tang et al. 2012).
In adaptive immune system, antigens can only be recognized by T cells when they are bound with HLA class II and class I molecule (Zhang et al. 2005). Human Leukocyte Antigens (HLA) are highly polymorphic in nature in human and also have ability to present range of peptide epitopes on surface of cells for recognition by T cells (Sharma et al. 2014). The most promiscuous T cell epitopes can bind with number of HLA supertype alleles to cover different populations which lowers the chance of antigen escape related to antigenic drift or shift. The concept of HLA supertype has a profound role in the perceptive of T cell epitope assortment, disintegration and discrimination during T cell immune responses (Kangueane et al. 2005). The immune system has two antigen processing and presentation pathways viz. cytosolic pathway and endocytic pathway. Endogenous antigens are processed in the cytosolic pathway which is presented with HLA class I molecules and exogenous antigen are processed by an endocytic pathway which is presented by HLA class II molecules. Transporter associated with antigen processing (TAP) protein is involved in transport of cytosolic peptides into Rough Endoplasmic Reticulum (RER) to further bind with HLA class I molecules during cytosolic pathway. TAP protein is present in the membrane of RER which act as channel between cytosol and lumen of RER. Therefore peptides binding to HLA class I and TAP protein is crucial factor to initiate an immune response (Procko and Gaudet 2009; Gaudet and Wiley 2001).
Here we present sequence and structure based study of identified promiscuous HLA and cTAP binding T cell epitopes. 3D structure models were generated for epitopes and corresponding HLA allele by Pepstr and Modeller 9.10 respectively for their structure based study. The docking of the identified consensus peptide nanomer epitope with cTAP1 and favored HLA alleles with long-term objective of vaccine design of JEV. The docking study of epitope with cTAP1 comes as a cross check that our identified conserved epitope is indeed well bound and channeled by the cTAP1 cavity from cytoplasm to ER lumen for HLA class I antigen processing and presentation. Further, best promiscuous epitopes analyzed for their binding stability to respective HLA allele by NAMD–VMD molecular dynamics simulation.
Methodology
Genotype-III strain JE viruses are widely distributed in Asia and hence are commonly used for vaccines development which also provides cross protection against others JEV genotypes (Singh et al. 2015a). The complete genome and protein sequences of genotype III JEV strain (Accession no. ABU94628) were obtained from sequence database NCBI.
Identification of Promiscuous Conserved T Cell Epitopes
All non-structural and structural proteins of JEV were examined for identification of possible dominant HLA class I binding T cell epitopes using immunoinformatics tools. Propred I and IEDB-ANN (Immune Epitope Data Base- Artificial Neural Network) MHC class I binding prediction tool (Singh and Raghava 2003; Nielsen et al. 2003) were employed for binding analysis of all possible peptides. First screening of epitopes was done against 47 HLA alleles by propred I at 1–4 % threshold and than propred screened epitopes were cross checked against more specifically to five (A2, A3, A24, B7 and B15) class I HLA supertypes (total 20 HLA alleles) by IEDB-ANN algorithm to cover maximum population.
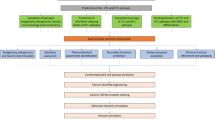
All the propred I predicted T cell epitopes of JEV were undergone for worldwide conservancy study among all five JEV genotypes by IEDB conservancy tool. To perform the conservancy analysis, all protein sequences of all genotypes belongs to different geographical regions were obtained randomly from NCBI database. The identified propred I nanomeric peptides which having less than 50 IEDB percentile value and 88–100 % conservancy with a maximum single mutation were selected for further study. Each identified and highly conserved epitopes were also analysed for their TAP binding property by MHC Pred 2.0 (Guan et al. 2006). This immunoinformatics top down approach helped in finding of promiscuous epitopes among identified all T cell epitopes (Fig. 1). Promiscuous epitopes are those epitope which bind with all HLA allele members of HLA supertypes (Burrows et al. 2003). Here the study includes 20 HLA alleles of five HLA class I supertypes viz. A2 (A*0201, A*0202, A*0203, A*0205, A*0206), A3 (A*0301, A*1101, A*3101, A*3301, A*6801), A24 (A*2402, B*3802), B7 (B*0702, B*3501, B*5101, B*5102, B*5301) and B15 (A*0101, B*1501 and B*1502) to cover maximum population (Reche and Reinherz 2005). Along with above analysis, the analysis included the two known antigenic peptide epitopes as positive controls, they were also analyzed for their IEDB percentile value against five (A2, A3, A24, B7 and B15) class I HLA supertype alleles to compare with identified epitopes. Hepatitis core protein (Accession number-CAA59535) peptide epitope 141STLPETTVV149 and H1N1 Nucleoprotein (Accession number-P03466) peptide epitope 265ILRGSVAHK273 are taken as positive control (Singh et al. 2015a; Ansari et al. 2009).
T Cell Epitopes Share the Part Sequence of B Cell Epitopes
For identification of those T cell epitopes which share sequence with longer fragments of B cell epitopes, all amino acid sequence of all proteins of JEV were subjected to BCPred. BCpred and AAP methods are used to predict the 20 amino acid fixed length epitope (Chen et al. 2007). Linear T cell and B cell epitopes on same peptide fragment is more achievable which can confer the protection by neutralizing antibody combined with an effective cell-mediated response. (Huber et al. 2015; Castelli et al. 2013) Finally the only those T cell epitope peptide sequences were preferred which were found in predicted B cell epitopes fragments for further structure based analysis.
3D Structural Modeling and Validation of Identified Epitopes and HLA Alleles
Among all identified epitopes, those T cell epitopes of JEV were chosen as potential epitopes on the basis of IEDB percentile score, high conservancy, population allele coverage, TAP binding and B cell epitope property for further structure based study. Epitopes were modeled by Pepstr (Kaur et al. 2007) and their structure also validated by Amber 6.0. All experimented HLA alleles sequence and their PDB structure were obtained from IMGT and PDB databases respectively (Robinson et al. 2015; Berman et al. 2000). Those HLA alleles structures were not retrieved from databases, will be modeled using corresponding templates by Modeller 9.10 (Sali 2014). HLA alleles models were further validated by using Errat (Colovos and Yeates 1993), ProSA (Wiederstein and Sippl 2007), ProQ (Wallner and Elofsson 2003) and RAMPAGE (Lovell et al. 2002).
Molecular Docking Study of Identified Epitopes with Their Favored Alleles and cTAP1
After structural modeling of selected epitopes and favored HLA alleles, molecular binding simulation was performed by Autodock 4.2 and Hex 8.0 (Morris et al. 2009; Ritchie et al. 2008). In docking experiments, water molecules were removed from receptor, added polar hydrogen to it and also added necessary charges (Gasteiger and Kollman charges etc.) to generate final docking.pdbqt file. In all docking experiments the ligands (epitopes) were kept free for bond rotation except peptide bonds. Discovery studio was used for analysis of the docked peptide and allele complexes. These binding analysis were further validated by Hex 8.0 interactive molecular graphics program. Hex 8.0, the fast fourier transformation based docking, we used grid dimension of 0.6 with range of 180 and step size 7.5. After binding analysis with favored HLA alleles, best HLA class I binding peptide was identified for docking study with cTAP1 to confirm their antigenic processing during cytosolic process for binding to HLA class I molecule.
Molecular Dynamic Simulation of Epitope-HLA Allele Complexes
NAnoscale Molecular Dynamics (NAMD) with Visual Molecular Dynamics (VMD) was used for Molecular dynamics (MD) simulation (James et al. 2005; Humphrey et al. 1996). In order to run MD simulation for epitope and allele complex, we generated a Protein complex Structure File (PSF) by accessing.pdb files through PSF builder tool of VMD. The file.psf was generated by using various force field parameters such as bond strengths, equilibrium lengths and various bonding interaction. Finally trajectory.dcd file generated by NAMD. Root mean square deviation (RMSD) value of the complex was calculated by using rmsd.tcl source file from the Tk console of VMD. Finally RMSD was saved as rmsd.dat file and Microsoft Excel was used to plot the values in the file rmsd.dat. RMSD graph was generated for an equilibrated MD simulation system of epitope and allele complexes.
Result
Identification of HLA Alleles, TAP Binding Epitopes and their Conservancy Study
Primary screening by propred I has identified total 102 HLA class I epitopes at 1–4 % threshold. Further these epitopes were cross checked by IEDB-ANN algorithm to select only those epitopes which having more than 8 or 10 HLA supertype allele binding frequency out of 20 HLA study alleles with less than 50 percentile IEDB score. Along with this analysis, two positive control known antigenic peptides of Hepatitis core protein (Accession number-CAA59535) peptide epitope 141STLPETTVV149 and H1N1 Nucleoprotein (Accession Number-P03466) peptide epitope 265ILRGSVAHK273 were also tested for IEDB-ANN algorithm to find their population coverage as shown in Table 1. Highly promiscuous ten epitopes were found highly conserved with high population coverage as shown in Table 1 supertype analysis with comparison to positive control antigenic peptides. Out of these ten T cell epitopes, two epitopes of glycoprotein (IPIVSVASL, LVTVNPFVA) were predicted most potential promiscuous epitopes in terms of HLA class I supertype allele binding frequency, TAP binding IC50 score, conserve nature and also share sequence with B cell epitope (Table 1). The epitope LVTVNPFVA was found 100 % conserve nature in all genotypes and also showed TAP binding property with IC50 value of 605.34 nM. But epitope IPIVSVASL showed 88 % conserve nature with single mutation (Valine replaced by Serine amino acid) IPIV(S G5 )SVASL in genotype V but remains conserved in rest all four JEV genotypes (I, II, III and IV). Findings revealed that LVTVNPFVA, IPIVSVASL epitopes binding to maximum members of A2, A3, A24, B7 and B15 HLA class I supertype with 18 and 16 HLA alleles frequency out of 20 HLA alleles, which is highest among all promiscuous and positive control epitopes as shown in Table 2. Positive control peptide epitopes 141STLPETTVV149 and 265ILRGSVAHK273 have shown 14 and 12 supertype alleles binding frequency out of total 20 supertype alleles with less than 50 IEDB percentile value in this population and supertype analysis respectively.
Therefore LVTVNPFVA, IPIVSVASL, epitopes were identified as super antigenic or most promiscuous and share sequence with 20 amino acid long fragments of B cell epitopes (Table 3). The T cell epitopes 55LVTVNPFVA63 and 38IPIVSVASL46 share sequence with fragment 51PVGRLVTVNPFVAASSANSK70 and 28SYSGSDGPCKIPIVSVASLN47 B cell epitopes of glycoprotein respectively. As reported, a protein fragment determined with linear B cell and T cell epitope is favorable to confer protective immunity and multiple epitopes if combined together can be basis of development of epitope vaccine (Castelli et al. 2013). These identified super antigenic epitopes and favored supertype HLA alleles, were modeled to analyze their binding simulations.
Structural Modeling of Identified Epitopes and HLA Alleles
Structural model of epitopes were generated by Pepstr, which were further refined with energy minimization and MD simulation using Amber 6.0. The IMGT/HLA Database and Protein Data Bank (PDB) Database allowed us to retrieve information upon a specific HLA allele sequences and their pdb structures. HLA class I, A*0101, A*0201, A*0301, B*0702, B*3501, B*5101 and B*5301 alleles and their PDB ID are 4NQV, 1A07, 3RL1, 3VCL, 1XH3, 1E27 and 1A10, were taken for docking experiments respectively. The B5102 allele structures was not retrieved from PDB database, will be modeled with the help of Modeller 9.10 by using template 1BII PDB. Structural model of allele B5102 was validated by using several tools viz. Errat, ProSA, Pro Q and RAMPAGE. Models quality was acceptable on the basis of OQF, LG score, Max Sub and Z score values (Fig. 2; Table 4). Residues in favored region were 91.6 % on Ramachandran plot for B5102 alleles as revealed by RAMPAGE.
Binding Simulation of Identified Epitopes and HLA Alleles
Docking study of identified epitopes LVTVNPFVA, IPIVSVASL with major HLA alleles of HLA class I supertypes were done to reveal their binding pattern. Binding of LVTVNPFVA and IPIVSVASL epitopes with B*5101 HLA allele showed best and favourable energy minimization of −5.70 and −5.46 kcal/mol, respectively. Stable complex of LVTVNPFVA-B5101 formed two H-bond viz. ASP310 and TYR 342 (Fig. 3). Similarly stable complex of IPIVSVASL-B*5101 allele formed three H bond viz. with ASP30, TYR302 and TYR339 (Fig. 4). Validation using Hex 8.0 also showed that LVTVNPFVA-B*5101 and IPIVSVASL-B*5101 interactions formed stable complexes with one H-bond. Binding energy of selected epitopes with favored HLA alleles by Autodock 4.2 and Hex 8.0, are tabulated in Tables 5 and 6.
Furthermore we also present the docking study of identified completely conserved epitope LVTVNPFVA with cTAP1 channel cavity facilitating the smooth passage of the epitope from cytoplasm to ER lumen. This docking study show that the epitope peptide gets hold at the upper part of the cavity by two hydrogen bonds and then with very optimal binding energy it is gripped by lower part of the cavity. If we put these two states of binding in sequence then we may conclude a smooth facilitation for epitope peptide transport from cytoplasm to ER lumen. The Docking study by Autodock 4.2 shows 2 best binding state in the cTAP1 cavity. In one binding state the epitope is forming 2 hydrogen bonds at the upper portion of cTAP1 viz. ARG515 and TYR555, having weak binding energy of −1.88 kcal/mol, while in other state which is in the lower cavity of cTAP1 forms no hydrogen bond and has very binding energy of −1.13 kcal/mol (Fig. 5).
The docking study of identified highly conserved epitope LVTVNPFVA (grey solid spheres) with cTAP1 (white solid spheres) channel cavity facilitating the smooth passage of the epitope from cytoplasm to ER lumen. a This docking study shows that the epitope peptide gets hold at the upper part of the cavity by two hydrogen bonds. b With very optimal binding energy it is gripped by lower part of the cavity. c The Docking study by Autodock 4.2 shows the epitope is forming 2 hydrogen bonds at the upper portion of cTAP1 viz. ARG515 and TYR555, having binding energy of −1.88 kcal/mol. If we put these two states of binding in sequence then we may conclude a smooth facilitation for peptide transport from cell cytoplasm to lumen of ER (Color figure online)
NAMD Simulation of Epitope-HLA Allele Complexes
The peptide and allele complexes formed by Autodock 4.2 were tested for their binding stability by NAMD-VMD simulation. Peptide allele complex (LVTVNPFVA-B5101) showed the highest RMSD value of 11.4 Å (Fig. 6). The RMSD values of the LVTVNPFVA-B*5101 allele complex are acceptable as shown a parallelism over the time window to conclude stable binding (Fig. 6).
Discussion
In the presented study, propred I and IEDB were employed for mapping of best T cell epitopes from proteome of JEV along with worldwide genotype conservancy. Only two LVTVNPFVA and IPIVSVASL T cell epitopes were identified as highly conserved, high HLA class I allele binding frequency, B cell epitope property and promiscuous with maximum population coverage among all predicted T cell epitopes along with two positive control peptides. Among two best epitopes, LVTVNPFVA epitope from glycoprotein of JEV was found as best binder in terms of the HLA allele coverage with 100 % conserve nature in all genotypes and was also a good cTAP1 binder in comparision to IPIVSVASL T cell epitope. TAP binding property is necessary to transport the cytosolic processed epitopes into lumen of RER through TAP protein to bind with MHC class I alleles. LVTVNPFVA peptide also show higher TAP binding IC50 score than IPIVSVASL peptide as mentioned in Table 1. Findings revealed that LVTVNPFVA, IPIVSVASL epitopes binding to maximum members of A2 (A*0201, A*0202, A*0203, A*0205, A*0206), A3(A*0301, A*1101, A*3101, A*3301, A*6801), A24 (A*2402, B*3802), B7 (B*0702, B*3501, B*5101, B*5102, B*5301) and B15 (A*0101, B*1501, B*1502) HLA class I supertype with less than 50 IEDB percentile value (Table 2). Therefore epitopes LVTVNPFVA and IPIVSVASL were identified as super antigenic or promiscuous peptide epitopes for JE.
Present immunoinformatics top down approach hence showed identified peptides are most promising vaccine candidates for JE, this was further validated by binding simulation analysis with favored HLA alleles of class I. Several other immunoinformatics top down approaches have given very promising results for finding of potential epitopes for viral diseases viz. H1N1 (Sharma and Kumar 2010) as well as tropical diseases viz. malaria, leshmaniasis (Singh et al. 2010, 2015b, c). Therefore our sequence based study results later validated by structure based study of most promiscuous T cell epitopes. The structure based study of epitope binding with HLA alleles revealed by Autodock 4.2 were further confirmed by Hex 8.0. The Hex energy of the LVTVNPFVA-B5101 complex obtained by Hex 8.0 was acceptable. Autodock 4.2 results for docking of LVTVNPFVA and B5101 revealed good binding energy of −5.70 kcal/mol and formed two H-bonds viz. ASP310 and TYR 342 amino acid residues. Negative free energy of binding is favourable for docking poses as per standard in docking methods (Morris et al. 2009). The NAMD-VMD study further confirms that the complex formed between the above mentioned peptide and allele indeed attains stable complex by showing parallalism with acceptable RMSD over a time window of 12,000 ps, with the highest peak at 11.4 Å. In support of this study, similar approach of immunoinformatics with structure based study was done for Toscana virus to find potential epitopes by getting good negative binding energy and good RMSD parallelism values during simulation for vaccine development (Jain et al. 2014). In addition to the above studies of the complex of epitope and alleles, we also performed the study of docking for epitope LVTVNPFVA with the cTAP1. This study revealed that the identified epitope is transported in a relay fashion through the cTAP1 cavity as shown in Fig. 5. The interaction of between epitope and cTAP is essential for antigen processing and presentation (Procko and Gaudet 2009; Gaudet and Wiley 2001). The docking results show cTAP1 channel cavity facilitating the smooth passage of the epitope from cytoplasm to ER lumen. The epitope gets hold at the upper part of the cavity by two hydrogen bonds and then with very optimal binding energy it is gripped by lower part of cavity. If we put these two states of binding in sequence then we may conclude a smooth facilitation for peptide transport from cytoplasm to ER lumen.
The present study has identified epitopes LVTVNPFVA and IPIVSVASL as most promising candidate for JE vaccine having super antigenic property. These identified potential novel peptide nanomer epitopes are more effective candidates in contrast to the vaccine candidates as whole viral proteins; furthermore it has been confirmed that few epitopes can represent immunogenicity of any protein (De Groot et al. 2002). Overall, as reported epitope based vaccines have shown good end result against H1N1, HIV, Tuberculosis, Malaria and Leshmaniasis diseases (Sharma and Kumar 2010; Jardine et al. 2013; Feng et al. 2013; Singh et al. 2010, 2015b) similarly the present study, identified epitopes could be tested as vaccine candidates and diagnostic reagents for JE as further prospects.
Conclusion
We document the identification of HLA class I binding JE viral peptides revealed from top down immunoinformatics approach. The finding reported here that the glycoprotein 55LVTVNPFVA63 and 38IPIVSVASL46 epitopes are highly conserved with super antigenic property and low binding energy values with B5101 allele. The identified epitope 55LVTVNPFVA63 of glycoprotein also form stable complex with B5101 allele; additionally smooth facilitation of epitope 55LVTVNPFVA63 through cTAP1 shows actual acceptance of the epitope in cytosolic antigen processing for further presentation by HLA class I. Hence these identified epitopes are most promising as vaccine candidates for JE and can also be useful as diagnostic agents for JE. Here we look forward to use these results as a platform for further trails for vaccine development against JE.
References
Ansari HR, Flower DR, Raghava GPS (2009) AntigenDB: an immunoinformatics database of pathogen antigens. Nucleic Acids Res 38:D847–D853
Barrett AD (1997) Japanese encephalitis and dengue vaccines. Biologicals 25:27–34
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acid Res 28(1):235–242
Burrows SR, Elkington RA, Miles JJ, Green KJ, Walker S, Haryana SM, Moss DJ, Dunckley H, Burrows JM, Khanna R (2003) Promiscuous CTL recognition of viral epitopes on multiple human leukocyte antigens: biological validation of the proposed HLA A24 supertype. J Immunol 171:1407–1412
Castelli M, Cappelletti F, Diotti RA, Sautto G, Criscuolo E, Peraro MD, Clementi N (2013) Peptide-Based vaccinology: experimental and computational approaches to target hypervariable viruses through the fine characterization of protective epitopes recognized by monoclonal antibodies and the identification of T-cell-activating peptides. Clin Dev Immunol. doi:10.1155/2013/521231
Chen J, Liu H, Yang J, Chou K (2007) Prediction of linear B-cell epitopes using amino acid pair antigenicity scale. Amino Acids 33:423–428
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of non-bonded atomic interactions. Protein Sci 2:1511–1519
De Groot AS, Sbai H, Aubin CS, McMurry J, Martin W (2002) Immuno-informatics: mining genomes for vaccine components. Immunol Cell Biol 80:255–269
Erra EO, Askling HH, Rombo L, Riutta J, Vene S, Yoksan S, Lindquist L, Pakkanen SH, Huhtamo E, Vapalahti O, Kantele A (2012) A single dose of vero cell-derived Japanese encephalitis (JE) vaccine (Ixiaro) effectively boosts immunity in travelers primed with mouse brain-derived JE vaccines. Clin Infect Dis 55:825–834
Erra EO, Askling HH, Yoksan S, Rombo L, Riutta J, Vene S, Lindquist L, Vapalahti O, Kantele A (2013) Cross-protection elicited by primary and booster vaccinations against Japanese encephalitis: a two-year follow-up study. Vaccine 32:119–123
Feng G, Jiang Q, Xia M, Lu Y, Qiu W, Zhao D, Lu L, Peng G, Wang Y (2013) Enhanced immune response and protective effects of Nano-chitosan-based DNA vaccine encoding T Cell epitopes of Esat-6 and FL against Mycobacterium tuberculosis infection. PLoS One 8(4):e61135
Gaudet R, Wiley DC (2001) Structure of the ABC ATPase domain of human TAP1, the transporter associated with antigen processing. EMBO J 20:4964–4972
Guan P, Hattotuwagama CK, Doytchinova IA, Flower DR (2006) MHCPred 2.0: an updated quantitative T-cell epitope prediction server. Appl Bioinform 5:55–61
Hoke CH, Vaughn DW, Nisalak A, Intralawan P, Poolsuppasit S, Jongsawas V, Titsyakorn U, Johnson RT (1992) Effect of high-dose dexamethasone on the outcome of acute encephalitis due to Japanese encephalitis virus. J Infect Dis 165:631–637
Huber SKR, Camps MGM, Jacobi RHJ, Mouthaan J, van Dijken JH, van Beek JF, Ossendorp JF, de Jonge JF (2015) Synthetic long peptide influenza vaccine containing conserved T and B Cell epitopes reduces viral load in lungs of mice and ferrets. PLoS One 10(6):e0127969
Humphrey W, Dalke A, Schulten K (1996) VMD—visual molecular dynamics. J Mol Gr 14:33–38
Igarashi A (2002) Control of Japanese encephalitis in Japan: immunization of humans and animals, and vector control. Curr Top Microbiol Immunol 267:139–152
Jain A, Tripathi P, Shrotiya A, Chaudhary R, Singh A (2014) In silico analysis and modeling of putative T cell epitopes for vaccine design of Toscana virus. 3 Biotech. doi:10.1007/s13205-014-0247-4
James CP, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26:1781–1802
Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, Nieusma T, Mathison J, Baker D, Ward AB, Burton DR, Stamatatos L, Nemazee D, Wilson IA, Schief WR (2013) Rational HIV immunogen design to target specific germline B cell receptors. Science 340(6133):711–716
Kangueane P, Sakharkar MK, Rajaseger G, Bolisetty S, Sivasekari B, Zhao B, Ravichandran M, Shapshak P, Subbiah S (2005) A framework to sub-type HLA supertypes. Front Biosci 10:879–886
Kaur H, Garg A, Raghava GPS (2007) PEPstr: a de novo method for tertiary structure prediction of small bioactive peptides. Protein Pept Lett 14:626–630
Lovell SC, Davis IW, Arendall WB, De Bakker P, Word JM, Prisant MG, Richardson JS, Richardson DC (2002) Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins 50:437–450
Monath TP (2002) Japanese encephalitis vaccines: current vaccines and future prospects. Curr Top Microbiol Immunol 267:105–138
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity. J. Comput Chem 16:2785–2791
Nielsen M, Lundegaard C, Worning P, Lauemøller SL, Lamberth K, Buus S, Brunak S, Lund O (2003) Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci 12(5):1007–1017
Okabe N (2005) Background of recent JE vaccine issues. Uirusu 55:303–306
Procko E, Gaudet R (2009) Antigen processing and presentation: tAPping into ABC transporters. Curr Opin Immunol 21:84–91
Reche PA, Reinherz EL (2005) PEPVAC: a web server for multi-epitope vaccine development based on the prediction of supertypic MHC ligands. Nucleic Acids Res 33:W138–W142
Ritchie DW, Kozakov D, Vajda S (2008) Accelerating Protein-Protein docking correlations using a six-dimensional analytic FFT generating function. Bioinform 24:1865–1873
Robinson J, Halliwell JA, Hayhurst JH, Flicek P, Parham P, Marsh SGE (2015) The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res 43:D423–D431
Sali WA (2014) Comparative protein structure modeling using modeller. Curr Protoc Bioinform. doi:10.1002/0471250953
Schiøler KL, Samuel M, Wai KL (2007) Vaccines for preventing Japanese encephalitis. Cochrane Database Syst Rev. doi:10.1002/14651858
Sharma P, Kumar A (2010) Immunoinformatics: screening of potential T cell antigenic determinants in proteome of H1N1 swine influenza virus for virus epitope vaccine design. J Proteom Bioinform 3:275–278
Sharma P, Saxena K, Mishra S, Kumar A (2014) A comprehensive analysis of predicted HLA binding peptides of JE viral proteins specific to north Indian isolates. Bioinformation 10:334–340
Shlim DR, Solomon T (2002) Japanese encephalitis vaccine for travelers: exploring the limits of risk. Clin Infect Dis 35:183–188
Singh SP, Mishra BN (2016) Major histocompatibility complex linked databases and prediction tools for designing vaccines. Hum Immunol 77:295–306
Singh H, Raghava GPS (2003) Propred I: prediction of HLA class-I binding sites. Bioinformatics 19:1009–1014
Singh SP, Khan F, Mishra BN (2010) Computational characterization of Plasmodium falciparum proteomic data for screening of potential vaccine candidates. Hum Immunol 71:136–143
Singh A, Mitra M, Sampath G, Venugopal P, Rao JV, Krishnamurthy B, Gupta MK, Sri Krishna S, Sudhakar B, Rao NB, Kaushik Y, Gopinathan K, Hegde NR, Gore MM, Krishna Mohan V, Ella KM (2015a) A Japanese encephalitis vaccine from India induces durable and cross-protective immunity against temporally and spatially wide-ranging global field strains. J Infect Dis 212(5):715–725
Singh SP, Roopendra K, Mishra BN (2015b) Genome-wide prediction of vaccine candidates for Leishmania major: an integrated approach. J Trop Med. doi:10.1155/2015/709216
Singh SP, Verma V, Mishra BN (2015c) Characterization of Plasmodium falciparum proteome at sexual blood stages for screening of effective vaccine candidates: an immunoinformatics approach. Immunol Immunog Insights 7:21–30
Tang H, Liu XS, Fang YZ, Pan L, Zhang ZW et al (2012) The epitopes of foot and mouth disease. Asian J Anim Vet Adv 7:1261–1265
Wallner B, Elofsson A (2003) Can correct protein models be identified. Protein Sci 12:1073–1086
WHO (2015) Weekly epidemiological record. WHO, Geneva, pp 69–88
Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35:407–410
Zhang GL, Khan AM, Srinivasan KN, August JT, Brusic V (2005) MULTIPRED: a computational system for prediction of promiscuous HLA binding peptides. Nucleic Acids Res 33:172–179
Acknowledgements
We are thankful to Prof. A. K. Ghosh, IFTM University, for his experimental guidance during the course of experimental work. We are also grateful to Institute of Engineering and Technology, Mangalayatan University, for providing necessary experimental facilities.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The authors declare that there were no animals or humans involved in this present study.
Rights and permissions
About this article
Cite this article
Sharma, P., Srivastav, S., Mishra, S. et al. Sequence and Structure Based Binding Prediction Study of HLA Class I and cTAP Binding Peptides for Japanese Encephalitis Vaccine Development. Int J Pept Res Ther 23, 269–279 (2017). https://doi.org/10.1007/s10989-016-9558-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-016-9558-0