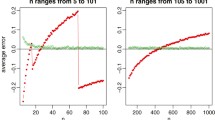
Abstract
Studies of chronic disease often involve modeling the relationship between marker processes and disease onset or progression. The Cox regression model is perhaps the most common and convenient approach to analysis in this setting. In most cohort studies, however, biospecimens and biomarker values are only measured intermittently (e.g. at clinic visits) so Cox models often treat biomarker values as fixed at their most recently observed values, until they are updated at the next visit. We consider the implications of this convention on the limiting values of regression coefficient estimators when the marker values themselves impact the intensity for clinic visits. A joint multistate model is described for the marker-failure-visit process which can be fitted to mitigate this bias and an expectation-maximization algorithm is developed. An application to data from a registry of patients with psoriatic arthritis is given for illustration.





Similar content being viewed by others
References
Aalen O, Borgan Ø, Fekjær H (2001) Covariate adjustment of event histories estimated from Markov chains: the additive approach. Biometrics 57(4):993–1001
Altman D (1991) Categorising continuous variables. Br J Cancer 64(5):975
Andersen P, Gill R (1982) Cox’s regression model for counting processes: a large sample study. The Annals of Statistics 10(4):1100–1120
Andersen P, Liestol K (2003) Attenuation caused by infrequently updated covariates in survival analysis. Biostatistics 4(4):633–649
Andy S, Keeffe E (2003) Elevated AST or ALT to nonalcoholic fatty liver disease: accurate predictor of disease prevalence? The American Journal of Gastroenterology 98(5):955–956
Butler A, English E, Kilpatrick E, Östlundh L, Chemaitelly H, Abu-Raddad L, Alberti K, Atkin S, John W (2021) Diagnosing type 2 diabetes using Hemoglobin A1c: a systematic review and meta-analysis of the diagnostic cutpoint based on microvascular complications. Acta Diabetologica 58(3):279–300
Cook R, Lawless J (2018) Multistate Models for the Analysis of Life History Data. Chapman and Hall/CRC, Boca Raton, FL
Cook R, Lawless J (2021) Independence conditions and the analysis of life history studies with intermittent observation. Biostatistics 22(3):455–481
Datta S, Satten G (2001) Validity of the Aalen-Johansen estimators of stage occupation probabilities and Nelson-Aalen estimators of integrated transition hazards for non-Markov models. Statistics & Probability Letters 55(4):403–411
de Bruijne M, Cessie S, Kluin-Nelemans H, Houwelingen H (2001) On the use of Cox regression in the presence of an irregularly observed time-dependent covariate. Stat Med 20(24):3817–3829
Dempster A, Laird N, Rubin D (1977) Maximum likelihood from incomplete data via the EM algorithm. J Royal Stat Soc: Series B (Methodological) 39(1):1–38
Eeg-Olofsson K, Cederholm J, Nilsson P, Zethelius B, Svensson AM, Gudbjörnsdottir S, Eliasson B (2010) New aspects of HbA1c as a risk factor for cardiovascular diseases in type 2 diabetes: an observational study from the Swedish National Diabetes Register (NDR). J Intern Med 268(5):471–482
Gelman A, Park D (2009) Splitting a predictor at the upper quarter or third and the lower quarter or third. The American Statistician 63(1):1–8
Gladman D, Chandran V (2011) Observational cohort studies: lessons learnt from the University of Toronto Psoriatic Arthritis Program. Rheumatology 50(1):25–31
Jewell N, Kalbfleisch J (1996) Marker processes in survival analysis. Lifetime Data Anal 2(1):15–29
Jiang S, Cook R, Zeng L (2020) Mitigating bias from intermittent measurement of time-dependent covariates in failure time analysis. Stat Med 39(13):1833–1845
Lin D, Wei LJ (1989) The robust inference for the Cox proportional hazards model. Journal of the American Statistical Association 84(408):1074–1078
Louis T (1982) Finding the observed information matrix when using the EM algorithm. J Royal Stat Soc: Series B (Methodological) 44(2):226–233
Martinussen T (1999) Cox regression with incomplete covariate measurements using the EM - algorithm. Scandinavian J Stat 26(4):479–491
McQuarrie E, Traynor J, Taylor A, Freel E, Fox J, Jardine A, Mark P (2014) Association between urinary sodium, creatinine, albumin, and long-term survival in chronic kidney disease. Hypertension 64(1):111–117
Papageorgiou G, Mauff K, Tomer A, Rizopoulos D (2019) An overview of joint modeling of time-to-event and longitudinal outcomes. Annual review of Statistics and its Application 6:223–240
R Core Team (2018) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria, https://www.R-project.org/
Raboud J, Reid N, Coates R, Farewell V (1993) Estimating risks of progressing to AIDS when covariates are measured with error. J Royal Stat Soc: Series A 156(3):393–406
Rahman P, Gladman D, Cook R, Zhou Y, Young G, Salonen D (1998) Radiological assessment in psoriatic arthritis. Br J Rheumatol 37(7):760–765
Royston P, Altman D, Sauerbrei W (2006) Dichotomizing continuous predictors in multiple regression: a bad idea. Stat Med 25(1):127–141
Struthers C, Kalbfleisch J (1986) Misspecified proportional hazard models. Biometrika 73(2):363–369
Tsiatis A, Davidian M (2001) A semiparametric estimator for the proportional hazards model with longitudinal covariates measured with error. Biometrika 88(2):447–458
Tsiatis A, Davidian M (2004) Joint modeling of longitudinal and time-to-event data: an overview. Statistica Sinica 14(3):809–834
Wong G, Chan H, Tse YK, Yip T, Lam K, Lui G, Wong V (2018) Normal on-treatment ALT during antiviral treatment is associated with a lower risk of hepatic events in patients with chronic hepatitis B. J Hepatol 69(4):793–802
Wulfsohn M, Tsiatis A (1997) A joint model for survival and longitudinal data measured with error. Biometrics 53(1):330–339
Acknowledgements
It is our great pleasure to contribute to the special issue of Lifetime Data Analysis in honour of David Oakes, whose many important contributions to the field of life history analysis are marked by their relevance, creativity, rigour, and clarity. We thank the Guest Editors Jong H. Jeong and Amita Manatunga for the opportunity to take part in this special and much-deserved recognition. This work was funded by the Natural Sciences and Engineering Research Council of Canada (RGPIN-2017-04207 for R.J.C. and RGPIN-2017-04055 for J.F.L.). R.J.C. is a Mathematics Faculty Research Chair at the University of Waterloo. The authors thank Drs. Dafna Gladman and Vinod Chandra of the Centre for Prognosis in Rheumatic Diseases for stimulating discussion and permission to use the data in the application.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Appendix
1.1 Computation of \(r^{(k)}(s)= E( {\bar{Y}}_i(s) d N_i(s))\)
Here we next describe the computation of the expectation \(r^{(0)}(s)= E\{ {\bar{Y}}_i(s) d N_i(s)\}\) based on this joint model. We use E to denote expectations taken with respect to the joint model depicted in Fig. 3 and P to denote corresponding probabilities based on the joint process. Note that
which can be computed as
If \(k=1\) we partition \(r^{(1)}(s) = E \{ {\bar{Y}}_i(s) X_i^{\circ }(s) d N_i(s) \}\) conformably with \(X^\circ (t) = (X^\circ _1(t), X_2')'\) and write \(r^{(1)}(s) = (r_1^{(1)}(s), [r^{(1)}_2(s)]')'\), the leading term has the form
This can be written as
since \({\bar{Y}}(s) X^{\circ }(s) = 1\) if and only if \({{\mathcal {Z}}}(s) \in {{\mathcal {C}}}^{zx^\circ }_{01} \cup {{\mathcal {C}}}^{zx^\circ }_{11}\). For \(r_2^{(1)}(s)\) we have
1.2 Computation of \(r^{(k)}(s;\psi )\)
Note that \(r^{(0)}(s; \psi ) = E \{ {\bar{Y}}_i(s) \exp (\psi _1 X_1^{\circ }(s) + \psi _2' X_2) \}\) is
which is computed as
We again partition \(r^{(1)}(s; \psi ) = E \{ {\bar{Y}}(s) X^{\circ }(s) \exp (\psi ' X^{\circ }(t) \} \) and note for the first element
The remaining \(p\times 1\) vector \(r_2^{(1)}(s;\psi )\) is given by
Together equations (A.1)–(A.6) facilitate computation of (10) and hence determination of \(\psi ^*\).
Joint model fitting via an EM algorithm
In what follows we give the complete data partial log-likelihoods and score functions for the intensities governing transitions in Fig. 3 and use \({{\mathcal {L}}}\) to distinguish these likelihoods from the observed data log-likelihood. The complete data here is conceived as containing the times of all marker transitions over (0, V), in which case the complete data likelihood can be factored into component functions that can be maximized separately (Cook and Lawless 2018, Chapter 2).
1.1 Complete data scores for the marker process
Markov intensities are adopted for the marker process giving the complete data partial log-likelihood contributions of the form
with \(d \varLambda _j(t | X_2) = d \varLambda _{jo}(t) \exp (\gamma '_j X_2)\) where \(d \varLambda _{jo}(t)\) is the baseline transition rate. Flexibility is desired for the marker process but semiparametric methods are challenging since the \(j \rightarrow 3-j\) transitions are not observed – we adopt piecewise-constant baseline rate functions and let \(0 = b_{j0}< b_{j1}< \cdots < b_{j, K_j - 1} = \infty \) denote cutpoints for the \(j \rightarrow j-1\) baseline rate, giving \({{\mathcal {B}}}_{jk} = [b_{j, k-1}, b_{jk})\), \(k = 1, \ldots , K_j\), \(j = 0, 1\). We let \(d \varLambda _{jo}(s)/ds = \exp (\alpha _{jk})\) if \(s \in {{\mathcal {B}}}_{jk}\) and \(\alpha _j = (\alpha _{j1}, \ldots , \alpha _{j K_j})'\). The complete data score function for the \(\alpha _{jk}\) are
which can be rewritten as
where \(N_{ijk} = \int _0^{\infty } I(s \in {{\mathcal {B}}}_{jk}) d {\bar{N}}_{ij}(s)\) is the number of \(j \rightarrow 1-j\) transitions over \({{\mathcal {B}}}_{jk}\) for indivdual i and \(S_{ijk} = \int _0^{\infty } {\bar{Y}}_{ij}(s) I(s \in {{\mathcal {B}}}_{jk}) ds\) is their time at risk for a transition out of state j in sub-interval \({{\mathcal {B}}}_{jk}\), \(j=0, 1\). The complete data partial score for \(\gamma _j\) is
and we write \(U_j = (U_{j1}, \ldots , U_{j K_j}, U'_{j, K_j + 1})'\) which is the estimating function for \(\theta _j = (\alpha '_j, \gamma '_j)'\). The key elements missing from the intermittent visit process are \(S_{ij} = (S_{ij1}, \ldots , S_{ijK_j})'\) and \(N_{ij} = (N_{ij1}, \ldots , N_{ijK_j})'\), \(j = 0, 1\).
1.2 Complete data scores for failure and censoring intensities
Here the complete data partial log-likelihood for the failure \((l=2)\) and censoring \((l=3)\) process intensities given by
We let \(0 = b_{l0}< b_{l1}< \cdots< b_{l, K_l - 1} < b_{l K_l} = \infty \) be \(K_l - 1\) cutpoints, giving intervals \({{\mathcal {B}}}_{lk}= (b_{l,k-1}, b_{lk})\) for the piecewise-constant intensities of the form \(d \varLambda _{lo}(t)/dt = \exp (\alpha _{lk})\) if \(t \in {{\mathcal {B}}}_{lk}\), \(k = 1, \ldots , K_l\) where \(d \varLambda _l(t | X(t)) = d \varLambda _{lo}(t) \exp (\beta ' X(t))\) if \(l=2\) and \(d \varLambda _l(t | X(t)) = d \varLambda _{lo}(t) \exp (\eta _c' X(t))\) if \(l=3\). The complete data score function for \(d\varLambda _{lo}(s)\) is
which under the piecewise-constant hazard model gives \(U_{lk} = \partial \log {{\mathcal {L}}}_l / \partial \alpha _{lk}\) of the form
where \(\phi = (\phi _1,\phi _2')'= \beta \) for \(l=2\) and \(\eta _c\) for \(l=3\). \({{\mathcal {S}}}_{ijk}^{(l)} = \int _0^{\infty } {\bar{Y}}_{ij}(s) I(s \in {{\mathcal {B}}}_{lk}) ds\) where the superscript (l) reflects the fact that this is the time at risk of a transition out of state j in interval \({{\mathcal {B}}}_{lk}\) (as opposed to \({{\mathcal {B}}}_{jk}\), \(j = 0, 1\)). For \(\phi \) we have \(U_{l, K_l + 1} = \partial \log {{\mathcal {L}}}_l / \partial \phi \)
1.3 Complete data scores for modulated Poisson visit process intensities
Here we let \(d \varLambda _4(t | X_i(t))\) be the rate function for visits giving a complete data partial log-likelihood for the visits intensity
For the visit process, we have \(0 = b_{40}< b_{41}< \cdots< b_{4, K_4 - 1} < b_{4 K_4} = \infty \) be \(K_4 - 1\) cutpoints, giving intervals \({{\mathcal {B}}}_{4k}= (b_{4,k-1},b_{4k})\), and let \(d \varLambda _{4o}(t)/dt = \exp (\alpha _{4k})\) if \(t \in {{\mathcal {B}}}_{4k}\), \(k = 1, \ldots , K_4\) where \(d \varLambda _4(t | X(t)) = d \varLambda _{4o}(t) \exp (\eta _a' X(t))\). The complete data score functions are \(U_{4k} = \partial \log {{\mathcal {L}}}_4 / \partial \alpha _{4k}\) and \(U_{4, K_4 + 1} = \partial \log {{\mathcal {L}}}_4 / \partial \eta _a\) given by
and
1.4 Computation of the conditional expectations
The elements that are missing include \(S_{ijk}\) and \(N_{ijk}\) in the complete data estimating functions for marker processes, and the covariate path \(\{X_i(u), 0 < u \}\) for the failure, censoring and visit process intensities. We let
denote the observed data for individual i, \(i = 1, \ldots , n\). Then
and
For the other estimating functions we require only
All of these expectations are based on the conditional probability \(P({{\mathcal {Z}}}_i(s) \in {{\mathcal {C}}}_j^z | D_i)\). We discuss how to compute this here by considering two special cases.
1.5 Case 1: \(a_{i r_i(u)}< u < a_{i, r_i(u) + 1}\) where \(A_i(u) = r_i(u)\)
The numerator of \(P({{\mathcal {Z}}}_i(u) \in {{\mathcal {R}}}_1^z | D_i)\) is given by
where \(z_i(u) = (r_i(u), 1, X_i^{\circ }(u))\) and the denominator is given in (12).
1.6 Case 2: \(a_{i r_i}< u < V_i\)
Here the numerator of \(P({{\mathcal {Z}}}_i(u) \in {{\mathcal {R}}}_1^z | D_i)\) is given below:
where \(z_i(u) = (r_i(u), 1, X_i^{\circ }(u))\) and the denominator is given by the observed data likelihood (12).
Rights and permissions
About this article
Cite this article
Cook, R.J., Lawless, J.F. & Xie, B. Marker-dependent observation and carry-forward of internal covariates in Cox regression. Lifetime Data Anal 28, 560–584 (2022). https://doi.org/10.1007/s10985-022-09561-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10985-022-09561-9