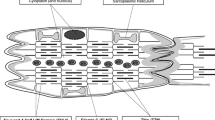
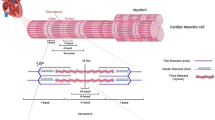
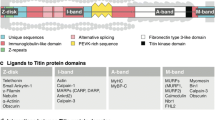
Abstract
Familial cardiomyopathy is an inherited disease that affects the structure and function of heart muscle and has an extreme range of phenotypes. Among the millions of affected individuals, patients with hypertrophic (HCM), dilated (DCM), or left ventricular non-compaction (LVNC) cardiomyopathy can experience morphologic changes of the heart which lead to sudden death in the most detrimental cases. TNNC1, the gene that codes for cardiac troponin C (cTnC), is a sarcomere gene associated with cardiomyopathies in which probands exhibit young age of presentation and high death, transplant or ventricular fibrillation events relative to TNNT2 and TNNI3 probands. Using GnomAD, ClinVar, UniProt and PhosphoSitePlus databases and published literature, an extensive list to date of identified genetic variants in TNNC1 and post-translational modifications (PTMs) in cTnC was compiled. Additionally, a recent cryo–EM structure of the cardiac thin filament regulatory unit was used to localize each functionally studied amino acid variant and each PTM (acetylation, glycation, s-nitrosylation, phosphorylation) in the structure of cTnC. TNNC1 has a large number of variants (> 100) relative to other genes of the same transcript size. Surprisingly, the mapped variant amino acids and PTMs are distributed throughout the cTnC structure. While many cardiomyopathy-associated variants are localized in α-helical regions of cTnC, this was not statistically significant χ2 (p = 0.72). Exploring the variants in TNNC1 and PTMs of cTnC in the contexts of cardiomyopathy association, physiological modulation and potential non-canonical roles provides insights into the normal function of cTnC along with the many facets of TNNC1 as a cardiomyopathic gene.






Similar content being viewed by others
Data availability
Data are already available in public databases.
Code availability
Not applicable.
References
Ardito F, Giuliani M, Perrone D, Troiano G, Lo Muzio L (2017) The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (Review). Int J Mol Med 40:271–280. https://doi.org/10.3892/ijmm.2017.3036
Asumda FZ, Chase PB (2012) Nuclear cardiac troponin and tropomyosin are expressed early in cardiac differentiation of rat mesenchymal stem cells. Differentiation 83:106–115. https://doi.org/10.1016/j.diff.2011.10.002
Badr MA, Pinto JR, Davidson MW, Chase PB (2016) Fluorescent protein-based Ca2+ sensor reveals global, divalent cation-dependent conformational changes in cardiac troponin C. PLoS ONE 11:e0164222. https://doi.org/10.1371/journal.pone.0164222
Bergmann O et al (2009) Evidence for cardiomyocyte renewal in humans. Science 324:98–102. https://doi.org/10.1126/science.1164680
Biesiadecki BJ, Kobayashi T, Walker JS, John Solaro R, de Tombe PP (2007) The troponin C G159D mutation blunts myofilament desensitization induced by troponin I Ser23/24 phosphorylation. Circ Res 100:1486–1493. https://doi.org/10.1161/01.RES.0000267744.92677.7f
Brieler J, Breeden MA, Tucker J (2017) Cardiomyopathy: an overview. Am Fam Physician 96:640–646
KN Brown VS Pendela RR Diaz 2020 Restrictive (infiltrative) cardiomyopathy. In: StatPearls. Treasure Island (FL)
Budde H et al (2019) The newly detected myosin binding protein C/troponin interaction, function and thin filaments integrity affected by cTn mutants. J Muscle Res Cell Motil 40:240. https://doi.org/10.1007/s10974-019-09534-w
Carnevale A et al (2020) Genomic study of dilated cardiomyopathy in a group of Mexican patients using site-directed next generation sequencing. Mol Genet Genomic Med. https://doi.org/10.1002/mgg3.1504
Chase PB, Martyn DA, Hannon JD (1994a) Activation dependence and kinetics of force and stiffness inhibition by aluminiofluoride, a slowly dissociating analogue of inorganic phosphate, in chemically skinned fibres from rabbit psoas muscle. JMRCM 15:119–129
Chase PB, Martyn DA, Hannon JD (1994b) Isometric force redevelopment of skinned muscle fibers from rabbit with and without Ca2+. Biophys J 67:1994–2001. https://doi.org/10.1016/S0006-3495(94)80682-4
Chase PB, Szczypinski MP, Soto EP (2013) Nuclear tropomyosin and troponin in striated muscle: new roles in a new locale? J Muscle Res Cell Motil 34:275–284. https://doi.org/10.1007/s10974-013-9356-7
Choudhry S, Puri K, Denfield SW (2019) An update on pediatric cardiomyopathy. Curr Treat Options Cardiovasc Med 21:36. https://doi.org/10.1007/s11936-019-0739-y
Chung WK, Kitner C, Maron BJ (2011) Novel frameshift mutation in Troponin C (TNNC1) associated with hypertrophic cardiomyopathy and sudden death. Cardiol Young 21:345–348. https://doi.org/10.1017/S1047951110001927
Costabel U, Wessendorf TE, Bonella F (2017) Epidemiology and clinical presentation of sarcoidosis. Klin Monbl Augenheilkd 234:790–795. https://doi.org/10.1055/s-0042-105569
Darling AL, Uversky VN (2018) Intrinsic disorder and posttranslational modifications: the darker side of the biological dark matter. Front Genet 9:158. https://doi.org/10.3389/fgene.2018.00158
Dong WJ, Jayasundar JJ, An J, Xing J, Cheung HC (2007) Effects of PKA phosphorylation of cardiac troponin I and strong crossbridge on conformational transitions of the N-domain of cardiac troponin C in regulated thin filaments. Biochemistry 46:9752–9761. https://doi.org/10.1021/bi700574n
Duan G, Walther D (2015) The roles of post-translational modifications in the context of protein interaction networks. PLoS Comput Biol. https://doi.org/10.1371/journal.pcbi.1004049
Dweck D, Hus N, Potter JD (2008) Challenging current paradigms related to cardiomyopathies. Are changes in the Ca2+ sensitivity of myofilaments containing cardiac troponin C mutations (G159D and L29Q) good predictors of the phenotypic outcomes? J Biol Chem 283:33119–33128. https://doi.org/10.1074/jbc.M804070200
Dweck D, Reynaldo DP, Pinto JR, Potter JD (2010) A dilated cardiomyopathy troponin C mutation lowers contractile force by reducing strong myosin-actin binding. J Biol Chem 285:17371–17379. https://doi.org/10.1074/jbc.M109.064105
Elliott PM et al (2014) 2014 ESC guidelines on diagnosis and management of hypertrophic cardiomyopathy: the Task Force for the Diagnosis and Management of Hypertrophic Cardiomyopathy of the European Society of Cardiology (ESC). Eur Heart J 35:2733–2779. https://doi.org/10.1093/eurheartj/ehu284
Figueiredo-Freitas C et al (2015) S-Nitrosylation of sarcomeric proteins depresses myofilament Ca2+ sensitivity in intact cardiomyocytes. Antioxid Redox Signal 23:1017–1034. https://doi.org/10.1089/ars.2015.6275
Franaszczyk M et al (2020) Analysis of de novo mutations in sporadic cardiomyopathies emphasizes their clinical relevance and points to novel candidate genes. J Clin Med. https://doi.org/10.3390/jcm9020370
Friedrich FW, Flenner F, Nasib M, Eschenhagen T, Carrier L (2016) Epigallocatechin-3-gallate accelerates relaxation and Ca2+ transient decay and desensitizes myofilaments in healthy and Mybpc3-targeted knock-in cardiomyopathic mice. Front Physiol 7:607. https://doi.org/10.3389/fphys.2016.00607
Fuchs F, Grabarek Z (2011) The Ca2+/Mg2+ sites of troponin C modulate crossbridge-mediated thin filament activation in cardiac myofibrils. Biochem Biophys Res Commun 408:697–700. https://doi.org/10.1016/j.bbrc.2011.04.092
Garfinkel AC, Seidman JG, Seidman CE (2018) Genetic pathogenesis of hypertrophic and dilated cardiomyopathy. Heart Fail Clin 14:139–146. https://doi.org/10.1016/j.hfc.2017.12.004
Gersh BJ et al (2011a) 2011 ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation 124:e783-831. https://doi.org/10.1161/CIR.0b013e318223e2bd
Gersh BJ et al (2011b) 2011 ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: executive summary: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation 124:2761–2796. https://doi.org/10.1161/CIR.0b013e318223e230
Gomes AV, Potter JD (2004) Molecular and cellular aspects of troponin cardiomyopathies. Ann N Y Acad Sci 1015:214–224
Gonzalez-Martinez D et al (2018) Structural and functional impact of troponin C-mediated Ca2+ sensitization on myofilament lattice spacing and cross-bridge mechanics in mouse cardiac muscle. J Mol Cell Cardiol 123:26–37. https://doi.org/10.1016/j.yjmcc.2018.08.015
Gordon AM, Homsher E, Regnier M (2000) Regulation of contraction in striated muscle. Physiol Rev 80:853–924. https://doi.org/10.1152/physrev.2000.80.2.853
Grabarek Z, Mabuchi Y, Gergely J (1995) Properties of troponin C acetylated at lysine residues. Biochemistry 34:11872–11881. https://doi.org/10.1021/bi00037a027
Grabarek Z, Tan R-Y, Wang J, Tao T, Gergely J (1990) Inhibition of mutant troponin C activity by an intra-domain disulphide bond. Nature 345:132–135. https://doi.org/10.1038/345132a0
Gupta MP, Samant SA, Smith SH, Shroff SG (2008) HDAC4 and PCAF bind to cardiac sarcomeres and play a role in regulating myofilament contractile activity. J Biol Chem 283:10135–10146. https://doi.org/10.1074/jbc.M710277200
Hannon JD, Chase PB, Martyn DA, Huntsman LL, Kushmerick MJ, Gordon AM (1993) Calcium-independent activation of skeletal muscle fibers by a modified form of cardiac troponin C. Biophys J 64:1632–1637
Harmel R, Fiedler D (2018) Features and regulation of non-enzymatic post-translational modifications. Nat Chem Biol 14:244–252. https://doi.org/10.1038/nchembio.2575
Harney DF, Butler RK, Edwards RJ (2005) Tyrosine phosphorylation of myosin heavy chain during skeletal muscle differentiation: an integrated bioinformatics approach. Theor Biol Med Model 2:12. https://doi.org/10.1186/1742-4682-2-12
Hershberger RE, Hedges DJ, Morales A (2013) Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol 10:531–547. https://doi.org/10.1038/nrcardio.2013.105
Hoffmann B, Schmidt-Traub H, Perrot A, Osterziel KJ, Geßner R (2001) First mutation in cardiac troponin C, L29Q, in a patient with hypertrophic cardiomyopathy. Hum Mutat 17:524. https://doi.org/10.1002/humu.1143
Huttlin EL et al (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143:1174–1189. https://doi.org/10.1016/j.cell.2010.12.001
Ikeda U, Minamisawa M, Koyama J (2015) Isolated left ventricular non-compaction cardiomyopathy in adults. J Cardiol 65:91–97. https://doi.org/10.1016/j.jjcc.2014.10.005
Ingles J et al (2019) Evaluating the clinical validity of hypertrophic cardiomyopathy genes. Circ Genom Precis Med 12:e002460. https://doi.org/10.1161/CIRCGEN.119.002460
Irie T et al (2015) S-nitrosylation of calcium-handling proteins in cardiac adrenergic signaling and hypertrophy. Circ Res 117:793–803. https://doi.org/10.1161/CIRCRESAHA.115.307157
Janssens JV, Ma B, Brimble MA, Van Eyk JE, Delbridge LMD, Mellor KM (2018) Cardiac troponins may be irreversibly modified by glycation: novel potential mechanisms of cardiac performance modulation. Sci Rep 8:16084. https://doi.org/10.1038/s41598-018-33886-x
Jefferies JL, Towbin JA (2010) Dilated cardiomyopathy. Lancet 375:752–762. https://doi.org/10.1016/S0140-6736(09)62023-7
Jenni R, Oechslin EN, van der Loo B (2007) Isolated ventricular non-compaction of the myocardium in adults. Heart 93:11–15. https://doi.org/10.1136/hrt.2005.082271
Johnston JR, Chase PB, Pinto JR (2018) Troponin through the looking-glass: emerging roles beyond regulation of striated muscle contraction. Oncotarget 9:1461–1482. https://doi.org/10.18632/oncotarget.22879
Johnston JR et al (2019) The intrinsically disordered C terminus of troponin T binds to troponin C to modulate myocardial force generation. J Biol Chem 294:20054–20069. https://doi.org/10.1074/jbc.RA119.011177
Katrukha IA (2013) Human cardiac troponin complex Structure and functions. Biochemistry (Mosc) 78:1447–1465. https://doi.org/10.1134/S0006297913130063
Kawai M, Johnston JR, Karam T, Wang L, Singh RK, Pinto JR (2017) Myosin rod hypophosphorylation and CB kinetics in papillary muscles from a TnC-A8V KI mouse model. Biophys J 112:1726–1736. https://doi.org/10.1016/j.bpj.2017.02.045
Kirk R et al (2009) Outcome of pediatric patients with dilated cardiomyopathy listed for transplant: a multi-institutional study. J Heart Lung Transplant 28:1322–1328. https://doi.org/10.1016/j.healun.2009.05.027
Kobayashi T, Solaro RJ (2005) Calcium, thin filaments, and the integrative biology of cardiac contractility. Annu Rev Physiol 67:39–67. https://doi.org/10.1146/annurev.physiol.67.040403.114025
Landim-Vieira M et al (2020) Familial dilated cardiomyopathy associated with a novel combination of compound heterozygous TNNC1 variants (* equal contributions). Front Physiol 10:1612. https://doi.org/10.3389/fphys.2019.01612
Landim-Vieira M, Schipper JM, Pinto JR, Chase PB (2020) Cardiomyocyte nuclearity and ploidy: when is double trouble? J Muscle Res Cell Motil. https://doi.org/10.1007/s10974-019-09545-7
Landstrom AP et al (2008) Molecular and functional characterization of novel hypertrophic cardiomyopathy susceptibility mutations in TNNC1-encoded troponin C. J Mol Cell Cardiol 45:281–288. https://doi.org/10.1016/j.yjmcc.2008.05.003
Li MX et al (2018) The calcium sensitizer drug MCI-154 binds the structural C-terminal domain of cardiac troponin C. Biochem Biophys Rep 16:145–151. https://doi.org/10.1016/j.bbrep.2018.10.012
Lim CC et al (2008) A novel mutant cardiac troponin C disrupts molecular motions critical for calcium binding affinity and cardiomyocyte contractility. Biophys J 94:3577–3589. https://doi.org/10.1529/biophysj.107.112896
Lu C et al (2018) Molecular analysis of inherited cardiomyopathy using next generation semiconductor sequencing technologies. J Transl Med 16:241. https://doi.org/10.1186/s12967-018-1605-5
Luk A, Ahn E, Soor GS, Butany J (2009) Dilated cardiomyopathy: a review. J Clin Pathol 62:219–225. https://doi.org/10.1136/jcp.2008.060731
Lundby A et al (2012) Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep 2:419–431. https://doi.org/10.1016/j.celrep.2012.07.006
Manning EP, Guinto PJ, Tardiff JC (2012) Correlation of molecular and functional effects of mutations in cardiac troponin T linked to familial hypertrophic cardiomyopathy: an integrative in silico/in vitro approach. J Biol Chem 287:14515–14523. https://doi.org/10.1074/jbc.M111.257436
Maron BJ, Maron MS (2013) Hypertrophic cardiomyopathy. Lancet 381:242–255. https://doi.org/10.1016/S0140-6736(12)60397-3
Maron BJ, Maron MS, Semsarian C (2012) Genetics of hypertrophic cardiomyopathy after 20 years: clinical perspectives. J Am Coll Cardiol 60:705–715. https://doi.org/10.1016/j.jacc.2012.02.068
Maron BJ et al (2006) Contemporary definitions and classification of the cardiomyopathies: an American Heart Association scientific statement from the council on clinical cardiology, heart failure and transplantation committee; quality of care and outcomes research and functional genomics and translational biology interdisciplinary working groups; and council on epidemiology and prevention. Circulation 113:1807–1816. https://doi.org/10.1161/CIRCULATIONAHA.106.174287
Marques MdA, de Oliveira GAP (2016) Cardiac troponin and tropomyosin: structural and cellular perspectives to unveil the hypertrophic cardiomyopathy phenotype. Front Physiol 7:429. https://doi.org/10.3389/fphys.2016.00429
Marques MdA et al (2017) Allosteric transmission along a loosely structured backbone allows a cardiac troponin C mutant to function with only one Ca2+ ion. J Biol Chem 292:2379–2394. https://doi.org/10.1074/jbc.M116.765362
Martins AS et al (2015) In vivo analysis of troponin C Knock-In (A8V) mice: evidence that TNNC1 is a hypertrophic cardiomyopathy susceptibility gene. Circ Cardiovasc Genet 8:653–664. https://doi.org/10.1161/CIRCGENETICS.114.000957
Martyn DA, Chase PB, Hannon JD, Huntsman LL, Kushmerick MJ, Gordon AM (1994) Unloaded shortening of skinned muscle fibers from rabbit activated with and without Ca2+. Biophys J 67:1984–1993. https://doi.org/10.1016/S0006-3495(94)80681-2
Mazzarotto F et al (2020) Reevaluating the genetic contribution of monogenic dilated cardiomyopathy. Circulation 141:387–398. https://doi.org/10.1161/CIRCULATIONAHA.119.037661
McKillop DFA, Geeves MA (1993) Regulation of the interaction between actin and myosin subfragment 1: evidence for three states of the thin filament. Biophys J 65:693–701. https://doi.org/10.1016/S0006-3495(93)81110-X
Miszalski-Jamka K et al (2017) Novel genetic triggers and genotype-phenotype correlations in patients with left ventricular noncompaction. Circ Cardiovasc Genet. https://doi.org/10.1161/CIRCGENETICS.117.001763
Mogensen J et al (2004) Severe disease expression of cardiac troponin C and T mutations in patients with idiopathic dilated cardiomyopathy. J Am Coll Cardiol 44:2033–2040. https://doi.org/10.1016/j.jacc.2004.08.027
Muchtar E, Blauwet LA, Gertz MA (2017) Restrictive cardiomyopathy: genetics, pathogenesis, clinical manifestations, diagnosis, and therapy. Circ Res 121:819–837. https://doi.org/10.1161/CIRCRESAHA.117.310982
Müller MM (2018) Post-translational modifications of protein backbones: unique functions, mechanisms, and challenges. Biochemistry 57:177–185. https://doi.org/10.1021/acs.biochem.7b00861
Musunuru K et al (2020) Genetic testing for inherited cardiovascular diseases: a scientific statement from the American Heart Association. Circ Genom Precis Med 13:e000067. https://doi.org/10.1161/HCG.0000000000000067
Negele JC, Dotson DG, Liu W, Sweeney HL, Putkey JA (1992) Mutation of the high affinity calcium binding sites in cardiac troponin C. J Biol Chem 267:825–831
Oda T, Yanagisawa H, Wakabayashi T (2020) Cryo-EM structures of cardiac thin filaments reveal the 3D architecture of troponin. J Struct Biol. https://doi.org/10.1016/j.jsb.2020.107450
Parvatiyar MS, Landstrom AP, Figueiredo-Freitas C, Potter JD, Ackerman MJ, Pinto JR (2012) A mutation in TNNC1-encoded cardiac troponin C, TNNC1-A31S, predisposes to hypertrophic cardiomyopathy and ventricular fibrillation. J Biol Chem 287:31845–31855. https://doi.org/10.1074/jbc.M112.377713
Pinto JR, Parvatiyar MS, Jones MA, Liang J, Ackerman MJ, Potter JD (2009) A functional and structural study of troponin C mutations related to hypertrophic cardiomyopathy. J Biol Chem 284:19090–19100. https://doi.org/10.1074/jbc.M109.007021
Pinto JR et al (2011) Functional characterization of TNNC1 rare variants identified in dilated cardiomyopathy. J Biol Chem 286:34404–34412. https://doi.org/10.1074/jbc.M111.267211
Pirani A, Xu C, Hatch V, Craig R, Tobacman LS, Lehman W (2005) Single particle analysis of relaxed and activated muscle thin filaments. J Mol Biol 346:761–772
Ploski R et al (2016) Evidence for troponin C (TNNC1) as a gene for autosomal recessive restrictive cardiomyopathy with fatal outcome in infancy. Am J Med Genet A 170:3241–3248. https://doi.org/10.1002/ajmg.a.37860
Poole KJV et al (2006) A comparison of muscle thin filament models obtained from electron microscopy reconstructions and low-angle X-ray fibre diagrams from non-overlap muscle. J Struct Biol 155:273–284. https://doi.org/10.1016/j.jsb.2006.02.020
Potter JD, Gergely J (1975) The calcium and magnesium binding sites on troponin and their role in the regulation of myofibrillar adenosine triphosphatase. J Biol Chem 250:4628–4633
Pua CJ et al (2020) Genetic studies of hypertrophic cardiomyopathy in singaporeans identify variants in TNNI3 and TNNT2 that are common in chinese patients. Circ Genom Precis Med. https://doi.org/10.1161/CIRCGEN.119.002823
Reichart D, Magnussen C, Zeller T, Blankenberg S (2019) Dilated cardiomyopathy: from epidemiologic to genetic phenotypes: a translational review of current literature. J Intern Med 286:362–372. https://doi.org/10.1111/joim.12944
Richards AA, Garg V (2010) Genetics of congenital heart disease. Curr Cardiol Rev 6:91–97. https://doi.org/10.2174/157340310791162703
Richards S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405–424. https://doi.org/10.1038/gim.2015.30
Risi C, Eisner J, Belknap B, Heeley DH, White HD, Schröder GF, Galkin VE (2017) Ca2+-induced movement of tropomyosin on native cardiac thin filaments revealed by cryoelectron microscopy. Proc Natl Acad Sci U S A 114:6782–6787. https://doi.org/10.1073/pnas.1700868114
Roher A, Leiska N, Spitz W (1986) The amino acid sequence of human cardiac troponin-C. Muscle Nerve 9:73–77. https://doi.org/10.1002/mus.880090112
Samsa LA, Yang B, Liu J (2013) Embryonic cardiac chamber maturation: trabeculation, conduction, and cardiomyocyte proliferation. Am J Med Genet C Semin Med Genet 163C:157–168. https://doi.org/10.1002/ajmg.c.31366
Schlecht W, Zhou Z, Li K-L, Rieck D, Ouyang Y, Dong W-J (2014) FRET study of the structural and kinetic effects of PKC phosphomimetic cardiac troponin T mutants on thin filament regulation. Arch Biochem Biophys 550–551:1–11. https://doi.org/10.1016/j.abb.2014.03.013
Schmidtmann A et al (2005) Cardiac troponin C-L29Q, related to hypertrophic cardiomyopathy, hinders the transduction of the protein kinase A dependent phosphorylation signal from cardiac troponin I to C. FEBS J 272:6087–6097. https://doi.org/10.1111/j.1742-4658.2005.05001.x
Semsarian C, Ingles J, Maron MS, Maron BJ (2015) New perspectives on the prevalence of hypertrophic cardiomyopathy. J Am Coll Cardiol 65:1249–1254. https://doi.org/10.1016/j.jacc.2015.01.019
Sia SK, Li MX, Spyracopoulos L, Gagné SM, Liu W, Putkey JA, Sykes BD (1997) Structure of cardiac muscle troponin C unexpectedly reveals a closed regulatory domain. J Biol Chem 272:18216–18221
DP Singh, H Patel 2020 Left Ventricular Non-compaction (LVNC) cardiomyopathy. In: StatPearls. Treasure Island (FL)
Slade DJ, Subramanian V, Fuhrmann J, Thompson PR (2014) Chemical and biological methods to detect post-translational modifications of arginine. Biopolymers 101:133–143. https://doi.org/10.1002/bip.22256
Slupsky CM, Sykes BD (1995) NMR solution structure of calcium-saturated skeletal muscle troponin C. Biochemistry 34:15953–15964
Solaro RJ, Moir AJ, Perry SV (1976) Phosphorylation of troponin I and the inotropic effect of adrenalin in the perfused rabbit heart. Nature 262:615–617
Swindle N, Tikunova SB (2010) Hypertrophic cardiomyopathy-linked mutation D145E drastically alters calcium binding by the C-domain of cardiac troponin C. Biochemistry 49:4813–4820. https://doi.org/10.1021/bi100400h
Tadros HJ et al (2020) Meta-analysis of cardiomyopathy-associated variants in troponin genes identifies loci and intragenic hot spots that are associated with worse clinical outcomes. J Mol Cell Cardiol 142:118–125. https://doi.org/10.1016/j.yjmcc.2020.04.005
Takasaki A et al (2018) Sarcomere gene variants act as a genetic trigger underlying the development of left ventricular noncompaction. Pediatr Res. https://doi.org/10.1038/s41390-018-0162-1
Takeda S, Yamashita A, Maeda K, Maéda Y (2003) Structure of the core domain of human cardiac troponin in the Ca2+-saturated form. Nature 424:35–41. https://doi.org/10.1038/nature01780
Tardiff JC (2011) Thin filament mutations: developing an integrative approach to a complex disorder. Circ Res 108:765–782. https://doi.org/10.1161/CIRCRESAHA.110.224170
Towbin JA et al (2006) Incidence, causes, and outcomes of dilated cardiomyopathy in children. JAMA 296:1867–1876. https://doi.org/10.1001/jama.296.15.1867
van der Velden J, Stienen GJM (2019) Cardiac disorders and pathophysiology of sarcomeric proteins. Physiol Rev 99:381–426. https://doi.org/10.1152/physrev.00040.2017
van Spaendonck-Zwarts KY et al (2010) Peripartum cardiomyopathy as a part of familial dilated cardiomyopathy. Circulation 121:2169–2175. https://doi.org/10.1161/CIRCULATIONAHA.109.929646
Vasilescu C et al (2018) Genetic basis of severe childhood-onset cardiomyopathies. J Am Coll Cardiol 72:2324–2338. https://doi.org/10.1016/j.jacc.2018.08.2171
Veltri T et al (2017) Hypertrophic cardiomyopathy cardiac troponin C mutations differentially affect slow skeletal and cardiac muscle regulation. Front Physiol 8:221. https://doi.org/10.3389/fphys.2017.00221
Vinogradova MV, Stone DB, Malanina GG, Karatzaferi C, Cooke R, Mendelson RA, Fletterick RJ (2005) Ca2+-regulated structural changes in troponin. Proc Natl Acad Sci U S A 102:5038–5043
Vos T et al (2016) Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 388:1545–1602. https://doi.org/10.1016/S0140-6736(16)31678-6
Wang X, Li MX, Sykes BD (2002) Structure of the regulatory N-domain of human cardiac troponin C in complex with human cardiac troponin I147–163 and bepridil. J Biol Chem 277:31124–31133. https://doi.org/10.1074/jbc.M203896200
Wang Y-C, Peterson SE, Loring JF (2014) Protein post-translational modifications and regulation of pluripotency in human stem cells. Cell Res 24:143–160. https://doi.org/10.1038/cr.2013.151
Warren CM et al (2015) Green tea catechin normalizes the enhanced Ca2+ sensitivity of myofilaments regulated by a hypertrophic cardiomyopathy-associated mutation in human cardiac troponin I (K206I). Circ Cardiovasc Genet 8:765–773. https://doi.org/10.1161/CIRCGENETICS.115.001234
Watkins WS et al (2019) De novo and recessive forms of congenital heart disease have distinct genetic and phenotypic landscapes. Nat Commun 10:4722. https://doi.org/10.1038/s41467-019-12582-y
Webber SA et al (2012) Outcomes of restrictive cardiomyopathy in childhood and the influence of phenotype: a report from the Pediatric Cardiomyopathy Registry. Circulation 126:1237–1244. https://doi.org/10.1161/CIRCULATIONAHA.112.104638
Weintraub RG, Semsarian C, Macdonald P (2017) Dilated cardiomyopathy. Lancet 390:400–414. https://doi.org/10.1016/S0140-6736(16)31713-5
Wilkinson JD et al (2010) The pediatric cardiomyopathy registry and heart failure: key results from the first 15 years. Heart Fail Clin 6(401–413):vii. https://doi.org/10.1016/j.hfc.2010.05.002
Willott RH, Gomes AV, Chang AN, Parvatiyar MS, Pinto JR, Potter JD (2010) Mutations in Troponin that cause HCM, DCM AND RCM: what can we learn about thin filament function? J Mol Cell Cardiol 48:882–892. https://doi.org/10.1016/j.yjmcc.2009.10.031
Wittekind SG, Ryan TD, Gao Z, Zafar F, Czosek RJ, Chin CW, Jefferies JL (2019) Contemporary outcomes of pediatric restrictive cardiomyopathy: a single-center experience. Pediatr Cardiol 40:694–704. https://doi.org/10.1007/s00246-018-2043-0
Wolf CM (2019) Hypertrophic cardiomyopathy: genetics and clinical perspectives. Cardiovasc Diagn Ther 9:S388–S415. https://doi.org/10.21037/cdt.2019.02.01
Wu H et al (2015) Epigenetic regulation of phosphodiesterases 2A and 3A underlies compromised β-adrenergic signaling in an iPSC model of dilated cardiomyopathy. Cell Stem Cell 17:89–100. https://doi.org/10.1016/j.stem.2015.04.020
Yamada Y, Namba K, Fujii T (2020) Cardiac muscle thin filament structures reveal calcium regulatory mechanism. Nat Commun 11:153. https://doi.org/10.1038/s41467-019-14008-1
Yang Y-S, Wang C-C, Chen B-H, Hou Y-H, Hung K-S, Mao Y-C (2015) Tyrosine sulfation as a protein post-translational modification. Molecules 20:2138–2164. https://doi.org/10.3390/molecules20022138
Yotti R, Seidman CE, Seidman JG (2019) Advances in the genetic basis and pathogenesis of sarcomere cardiomyopathies. Annu Rev Genomics Hum Genet 20:129–153. https://doi.org/10.1146/annurev-genom-083118-015306
Zhaolu WQ, Luo J, Pan B, Liu L-J, Tian J (2020) Cardiac troponin I R193H mutant interacts with HDAC1 to repress phosphodiesterase 4D expression in cardiomyocytes. Genes Dev. https://doi.org/10.1016/j.gendis.2020.01.004
Zhao W et al (2020) Epigenetic regulation of phosphodiesterase 4d in restrictive cardiomyopathy mice with cTnI mutations. Sci China Life Sci 63:563–570. https://doi.org/10.1007/s11427-018-9463-9
Zot HG, Hasbun JE, Michell CA, Landim-Vieira M, Pinto JR (2016) Enhanced troponin I binding explains the functional changes produced by the hypertrophic cardiomyopathy mutation A8V of cardiac troponin C. Arch Biochem Biophys 601:97–104. https://doi.org/10.1016/j.abb.2016.03.011
Acknowledgments
We thank Mark P. Szczypinski and Elliott P. Soto for excellent assistance in examining the predicted effects of single amino acid changes in cTnC on cellular localization.
Funding
This work was supported by U.S. American Heart Association Grant 19PRE34380628 (to J.R.J.), Florida State University’s Undergraduate Research Opportunity Program (UROP) (to P.B.C.), American Heart Association Scientist Development Grant 16SDG29120002 (to M.S.P.), Duke University School of Medicine (to A.P.L.) and U.S. National Institutes of Health National Heart Lung and Blood Institute grant HL128683 (to J.R.P.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Reinoso, T.R., Landim-Vieira, M., Shi, Y. et al. A comprehensive guide to genetic variants and post-translational modifications of cardiac troponin C. J Muscle Res Cell Motil 42, 323–342 (2021). https://doi.org/10.1007/s10974-020-09592-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10974-020-09592-5