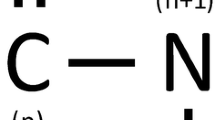Abstract
The results of secondary structure prediction methods are widely used in applications in biotechnology and bioinformatics. However, the accuracy limit of these methods could be improved up to 92%. One approach to achieve this goal is to harvest information from the primary structure of the peptide. This study aims to contribute to this goal by investigating the variations in propensity of amino acid pairings to α-helices in globular proteins depending on helix length. (n):(n + 4) residue pairings were determined using a comprehensive peptide data set according to backbone hydrogen bond criterion which states that backbone hydrogen bond is the dominant driving force of protein folding. Helix length is limited to 13 to 26 residues. Findings of this study show that propensities of ALA:GLY and GLY:GLU pairings to α-helix in globular protein increase with increasing helix length but of ALA:ALA and ALA:VAL decrease. While the frequencies of ILE:ALA, LEU:ALA, LEU:GLN, LEU:GLU, LEU:LEU, MET:ILE and VAL:LEU pairings remain roughly constant with length, the 25 residue pairings have varying propensities in narrow helix lengths. The remaining pairings have no prominent propensity to α-helices.





Similar content being viewed by others
Data Availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information Files).
References
Chou PY, Fasman GD (1974) Prediction of protein conformation. Biochemistry 13(2):222–245
Deleage G, Roux B (1987) An algorithm for protein secondary structure prediction based on class prediction. Protein Eng 1(4):289–294
Frishman D, Argos P (1995) Knowledge-based protein secondary structure assignment. Proteins 23(4):566–579
Garnier J, Gibrat JF, Robson B (1996) GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol 266:540–553
Geourjon C, Deleage G (1995) SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci 11(6):681–684
Gibrat JF, Garnier J, Robson B (1987) Further developments of protein secondary structure prediction using information theory. New parameters and consideration of residue pairs. J Mol Biol 198(3):425–443
Guermeur Y, Geourjon C, Gallinari P, Deleage G (1999) Improved performance in protein secondary structure prediction by inhomogeneous score combination. Bioinformatics 15(5):413–421
King RD, Sternberg MJ (1996) Identification and application of the concepts important for accurate and reliable protein secondary structure prediction. Protein Sci 5(11):2298–2310
Levin JM, Robson B, Garnier J (1986) An algorithm for secondary structure determination in proteins based on sequence similarity. FEBS Lett 205(2):303–308
Rost B, Sander C (1994) Combining evolutionary information and neural networks to predict protein secondary structure. Proteins 19(1):55–72
Garnier J, Osguthorpe DJ, Robson B (1978) Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol 120(1):97–120
Drozdetskiy A, Cole C, Procter J, Barton GJ (2015) JPred4: a protein secondary structure prediction server. Nucleic Acids Res 43(W1):W389–W394
Bernhofer M, Dallago C, Karl T, Satagopam V, Heinzinger M, Littmann M et al (2021) PredictProtein—predicting protein structure and function for 29 years. Nucleic Acids Res 49(W1):W535–W540
Jones DT (1999) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292(2):195–202
Zvelebil MJ, Barton GJ, Taylor WR, Sternberg MJ (1987) Prediction of protein secondary structure and active sites using the alignment of homologous sequences. J Mol Biol 195(4):957–961
Salamov AA, Solovyev VV (1995) Prediction of protein secondary structure by combining nearest-neighbor algorithms and multiple sequence alignments. J Mol Biol 247(1):11–15
Wako H, Blundell TL (1994) Use of amino acid environment-dependent substitution tables and conformational propensities in structure prediction from aligned sequences of homologous proteins. II. Secondary structures. J Mol Biol 238(5):693–708
Mehta PK, Heringa J, Argos P (1995) A simple and fast approach to prediction of protein secondary structure from multiply aligned sequences with accuracy above 70%. Protein Sci 4(12):2517–2525
Haber E, Anfinsen CB (1961) Regeneration of enzyme activity by air oxidation of reduced subtilisin-modified ribonuclease. J Biol Chem 236:422–424
Pirovano W, Heringa J (2010) Protein secondary structure prediction. Methods Mol Biol 609:327–348
Li Z, Yang Y, Zhan J, Dai L, Zhou Y (2013) Energy functions in de novo protein design: current challenges and future prospects. Annu Rev Biophys 42:315–335
Yang Y, Gao J, Wang J, Heffernan R, Hanson J, Paliwal K et al (2018) Sixty-five years of the long march in protein secondary structure prediction: the final stretch? Brief Bioinform 19(3):482–494
Ho CT, Huang YW, Chen TR, Lo CH, Lo WC (2021) Discovering the ultimate limits of protein secondary structure prediction. Biomolecules 11(11):1627
Wardah W, Khan MGM, Sharma A, Rashid MA (2019) Protein secondary structure prediction using neural networks and deep learning: a review. Comput Biol Chem 81:1–8
Rost B (2001) Review: protein secondary structure prediction continues to rise. J Struct Biol 134(2–3):204–218
de Sousa MM, Munteanu CR, Pazos A, Fonseca NA, Camacho R, Magalhaes AL (2011) Amino acid pair- and triplet-wise groupings in the interior of alpha-helical segments in proteins. J Theor Biol 271(1):136–144
Frishman D, Argos P (1996) Incorporation of non-local interactions in protein secondary structure prediction from the amino acid sequence. Protein Eng 9(2):133–142
Fonseca NA, Camacho R, Magalhaes AL (2008) Amino acid pairing at the N- and C-termini of helical segments in proteins. Proteins 70(1):188–196
Acevedo OE, Lareo LR (2005) Amino acid propensities revisited. OMICS 9(4):391–399
Chakrabartty A, Baldwin RL (1995) Stability of alpha-helices. Adv Protein Chem 46:141–176
Chakrabartty A, Kortemme T, Baldwin RL (1994) Helix propensities of the amino acids measured in alanine-based peptides without helix-stabilizing side-chain interactions. Protein Sci 3(5):843–852
Creamer TP, Rose GD (1992) Side-chain entropy opposes alpha-helix formation but rationalizes experimentally determined helix-forming propensities. Proc Natl Acad Sci USA 89(13):5937–5941
Engel DE, DeGrado WF (2004) Amino acid propensities are position-dependent throughout the length of alpha-helices. J Mol Biol 337(5):1195–1205
Horovitz A, Matthews JM, Fersht AR (1992) Alpha-helix stability in proteins. II. Factors that influence stability at an internal position. J Mol Biol 227(2):560–568
Pace CN, Scholtz JM (1998) A helix propensity scale based on experimental studies of peptides and proteins. Biophys J 75(1):422–427
Serrano L, Sancho J, Hirshberg M, Fersht AR (1992) Alpha-helix stability in proteins. I. Empirical correlations concerning substitution of side-chains at the N and C-caps and the replacement of alanine by glycine or serine at solvent-exposed surfaces. J Mol Biol 227(2):544–559
Sundaralingam M, Sekharudu YC, Yathindra N, Ravichandran V (1987) Ion-pairs in alpha-helices. Proteins Struct Funct Genet 2(1):64–71
Best RB, de Sancho D, Mittal J (2012) Residue-specific alpha-helix propensities from molecular simulation. Biophys J 102(6):1462–1467
Rohl CA, Fiori W, Baldwin RL (1999) Alanine is helix-stabilizing in both template-nucleated and standard peptide helices. Proc Natl Acad Sci USA 96(7):3682–3687
Nacar C (2020) Propensities of amino acid pairings in secondary structure of globular proteins. Protein J 39(1):21–32
Bruch MD, Dhingra MM, Gierasch LM (1991) Side chain-backbone hydrogen bonding contributes to helix stability in peptides derived from an alpha-helical region of carboxypeptidase A. Proteins 10(2):130–139
Presta LG, Rose GD (1988) Helix signals in proteins. Science 240(4859):1632–1641
Rose GD, Fleming PJ, Banavar JR, Maritan A (2006) A backbone-based theory of protein folding. Proc Natl Acad Sci USA 103(45):16623–16633
Wang J, Feng JA (2003) Exploring the sequence patterns in the alpha-helices of proteins. Protein Eng 16(11):799–807
Berman H, Henrick K, Nakamura H (2003) Announcing the worldwide Protein Data Bank. Nat Struct Biol 10(12):980
Baker EN, Hubbard RE (1984) Hydrogen bonding in globular proteins. Prog Biophys Mol Biol 44(2):97–179
Richardson JS, Richardson DC (1988) Amino acid preferences for specific locations at the ends of alpha helices. Science 240(4859):1648–1652
Serrano L, Fersht AR (1989) Capping and alpha-helix stability. Nature 342(6247):296–299
Kumar S, Bansal M (1996) Structural and sequence characteristics of long alpha helices in globular proteins. Biophys J 71(3):1574–1586
Makhatadze GI (2005) Thermodynamics of alpha-helix formation. Adv Protein Chem 72:199–226
Murphy KP, Freire E (1992) Thermodynamics of structural stability and cooperative folding behavior in proteins. Adv Protein Chem 43:313–361
Aurora R, Creamer TP, Srinivasan R, Rose GD (1997) Local interactions in protein folding: lessons from the alpha-helix. J Biol Chem 272(3):1413–1416
Munoz V, Serrano L (1995) Helix design, prediction and stability. Curr Opin Biotechnol 6(4):382–386
Baldwin RL (2003) In search of the energetic role of peptide hydrogen bonds. J Biol Chem 278(20):17581–17588
Guo H, Karplus M (1994) Solvent influence on the stability of the peptide hydrogen-bond—a supramolecular cooperative effect. J Phys Chem US 98(29):7104–7105
Ireta J, Neugebauer J, Scheffler M, Rojo A, Galvan M (2003) Density functional theory study of the cooperativity of hydrogen bonds in finite and infinite alpha-helices. J Phys Chem B 107(6):1432–1437
Rossi M, Scheffler M, Blum V (2013) Impact of vibrational entropy on the stability of unsolvated peptide helices with increasing length. J Phys Chem B 117(18):5574–5584
Tkatchenko A, Rossi M, Blum V, Ireta J, Scheffler M (2011) Unraveling the stability of polypeptide helices: critical role of van der Waals Interactions. Phys Rev Lett 106(11):118102
Wieczorek R, Dannenberg JJ (2004) Comparison of fully optimized alpha- and 3(10)-helices with extended beta-strands. An ONIOM density functional theory study. J Am Chem Soc 126(43):14198–14205
Funding
The authors did not receive support from any organization for the submitted work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Author declares that he has no conflicts of interest.
Ethical Approval
This article does not contain any studies with human or animal participants by the author.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nacar, C. Propensities of Some Amino Acid Pairings in α-Helices Vary with Length. Protein J 41, 551–562 (2022). https://doi.org/10.1007/s10930-022-10076-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-022-10076-3