Abstract
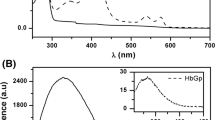
Biomacromolecules evolve and function inside the cell under crowded conditions. The effect of macromolecular crowding and confinement on nature and interactions of biomacromolecules cannot be ruled out. This study demonstrates the effect of volume exclusion due to macromolecular crowding on refolding rate of Gn-HCl induced unfolded hemoglobin. The in vivo like crowding milieu was created using dextran 70. Unfolding of Hb was followed by the absorbance at 280 nm and intrinsic fluorescence intensity along with a bathochromic shift that shows the destabilization of Hb in the presence of the denaturing agent. This was supported by a decrease in soret absorbance, increased hydrodynamic radii and loss in secondary structure, evidenced from dynamic light scattering and circular dichroism experiments respectively. Refolding process of Hb was followed by an increase in soret absorbance, decrease in intrinsic fluorescence intensity with a hypsochromic shift, decreased hydrodynamic radii and gain in secondary structural content. The results revealed that the effect of confinement and volume exclusion is insignificant on the process of Hb refolding.






Similar content being viewed by others
Data Availability
Not applicable.
Code Availability
Not applicable.
References
Siddiqui GA, Naeem A (2020) The contrasting effect of macromolecular crowding and confinement on fibril formation of globular protein: Underlying cause of proteopathies. J Mol Liq 322:114602
Fazili AN, Siddiqui AG, Bhat AS, et al (2015) Rifampicin induced aggregation of ovalbumin: malicious behaviour of antibiotics. Protein Pept Lett 22:644–653
Uversky VN, Karnoup AS, Segel DJ et al (1998) Anion-induced folding of Staphylococcal nuclease: characterization of multiple equilibrium partially folded intermediates. J Mol Biol 278:879–894
Muzammil S, Kumar Y, Tayyab S (2000) Anion-induced stabilization of human serum albumin prevents the formation of intermediate during urea denaturation. Proteins Struct Funct Bioinforma 40:29–38
Fink AL, Oberg KA, Seshadri S (1998) Discrete intermediates versus molten globule models for protein folding: characterization of partially folded intermediates of apomyoglobin. Fold Des 3:19–25
Ahmad B, Ahmed MZ, Haq SK, Khan RH (2005) Guanidine hydrochloride denaturation of human serum albumin originates by local unfolding of some stable loops in domain III. Biochim Biophys Acta (BBA) Proteins Proteomics 1750:93–102
Mirdha L, Chakraborty H (2020) Fluorescence quenching by ionic liquid as a potent tool to study protein unfolding intermediates. J Mol Liq 312:113408
Zhou H-X, Rivas G, Minton AP (2008) Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu Rev Biophys 37:375–397
Gnutt D, Ebbinghaus S (2016) The macromolecular crowding effect–from in vitro into the cell. Biol Chem 397:37–44
Silverstein TP, Slade K (2019) Effects of macromolecular crowding on biochemical systems. J Chem Educ 96:2476–2487
Minton AP, Wilf J (1981) Effect of macromolecular crowding upon the structure and function of an enzyme: glyceraldehyde-3-phosphate dehydrogenase. Biochemistry 20:4821–4826
Bhakuni K, Venkatesu P (2019) Does macromolecular crowding compatible with enzyme stem bromelain structure and stability? Int J Biol Macromol 131:527–535
Rajapaksha A, Stanley CB, Todd BA (2015) Effects of macromolecular crowding on the structure of a protein complex: a small-angle scattering study of superoxide dismutase. Biophys J 108:967–974
Fan Y-Q, Liu H-J, Li C et al (2012) Effects of macromolecular crowding on refolding of recombinant human brain-type creatine kinase. Int J Biol Macromol 51:113–118
Stagg L, Christiansen A, Wittung-Stafshede P (2011) Macromolecular crowding tunes folding landscape of parallel α/β protein, apoflavodoxin. J Am Chem Soc 133:646–648
Cook EC, Creamer TP (2016) Calcineurin in a crowded world. Biochemistry 55:3092–3101
Li J, Zheng H, Feng C (2019) Effect of macromolecular crowding on the FMN–Heme intraprotein electron transfer in inducible NO synthase. Biochemistry 58:3087–3096
Beg I, Minton AP, Islam A et al (2018) Comparison of the thermal stabilization of proteins by oligosaccharides and monosaccharide mixtures: Measurement and analysis in the context of excluded volume theory. Biophys Chem 237:31–37
Siddiqui GA, Naeem A (2018) Aggregation of globular protein as a consequences of macromolecular crowding: A time and concentration dependent study. Int J Biol Macromol 108:360–366. https://doi.org/10.1016/j.ijbiomac.2017.12.001
Li X, Zheng W, Zhang L et al (2009) Effective electrochemical method for investigation of hemoglobin unfolding based on the redox property of heme groups at glassy carbon electrodes. Anal Chem 81:8557–8563
Zhang G, Chen X, Guo J, Wang J (2009) Spectroscopic investigation of the interaction between chrysin and bovine serum albumin. J Mol Struct 921:346–351
Chi Z, Zhao J, You H, Wang M (2016) Study on the mechanism of interaction between phthalate acid esters and bovine hemoglobin. J Agric Food Chem 64:6035–6041
Yan Y-B, Wang Q, He H-W, Zhou H-M (2004) Protein thermal aggregation involves distinct regions: sequential events in the heat-induced unfolding and aggregation of hemoglobin. Biophys J 86:1682–1690
Kristinsson HG (2002) Acid-induced unfolding of flounder hemoglobin: evidence for a molten globular state with enhanced pro-oxidative activity. J Agric Food Chem 50:7669–7676
Mai Z, Zhao X, Dai Z, Zou X (2010) Contributions of components in guanidine hydrochloride to hemoglobin unfolding investigated by protein film electrochemistry. J Phys Chem B 114:7090–7097
Yamaguchi S, Yamamoto E, Mannen T et al (2013) Protein refolding using chemical refolding additives. Biotechnol J 8:17–31
Mach H, Volkin DB, Burke CJ, Middaugh CR (1995) Ultraviolet absorption spectroscopy. Protein Stab Fold 91–114
Street TO, Courtemanche N, Barrick D (2008) Protein folding and stability using denaturants. Methods Cell Biol 84:295–325
Togashi DM, Ryder AG, O’Shaughnessy D (2010) Monitoring local unfolding of bovine serum albumin during denaturation using steady-state and time-resolved fluorescence spectroscopy. J Fluoresc 20:441–452
Chen Y, Barkley MD (1998) Toward understanding tryptophan fluorescence in proteins. Biochemistry 37:9976–9982
Qiu W, Li T, Zhang L et al (2008) Ultrafast quenching of tryptophan fluorescence in proteins: Interresidue and intrahelical electron transfer. Chem Phys 350:154–164
Andrade SM, Costa SMB (2000) The location of tryptophan, N-acetyltryptophan and α‐chymotrypsin in reverse micelles of AOT: a fluorescence study¶. Photochem Photobiol 72:444–450
TAKE T (1961) On the dissociation of hemoglobin under the action of urea. J Biochem 49:206–210
Boys BL, Konermann L (2007) Folding and assembly of hemoglobin monitored by electrospray mass spectrometry using an on-line dialysis system. J Am Soc Mass Spectrom 18:8–16
Bhattacharya J, GhoshMoulick R, Choudhuri U et al (2004) Unfolding of hemoglobin variants—insights from urea gradient gel electrophoresis photon correlation spectroscopy and zeta potential measurements. Anal Chim Acta 522:207–214
Carvalho JWP, Santiago PS, Batista T et al (2012) On the temperature stability of extracellular hemoglobin of Glossoscolex paulistus, at different oxidation states: SAXS and DLS studies. Biophys Chem 163:44–55
Iram A, Alam T, Khan JM et al (2013) Molten globule of hemoglobin proceeds into aggregates and advanced glycated end products. PLoS One 8:e72075
Iram A, Naeem A (2013) Detection and analysis of protofibrils and fibrils of hemoglobin: implications for the pathogenesis and cure of heme loss related maladies. Arch Biochem Biophys 533:69–78
Siddiqui GA, Siddiqi MK, Khan RH, Naeem A (2018) Probing the binding of phenolic aldehyde vanillin with bovine serum albumin: Evidence from spectroscopic and docking approach. Spectrochim Acta Part A Mol Biomol Spectrosc 203:40–47
Christiansen A, Wang Q, Cheung MS and Wittung-Stafshede P (2013) Effects of macromolecular crowding agents on protein folding in vitro and in silico. Biophys Rev 5(2):137–145
Li J, Zhang S, Wang CC (2001) Effects of macromolecular crowding on the refolding of glucose-6-phosphate dehydrogenase and protein disulfide isomerase. J Biol Chem 276:34396–34401. https://doi.org/10.1074/jbc.M103392200
Zhou HX (2008) Effect of mixed macromolecular crowding agents on protein folding. Proteins Struct Funct Genet 72:1109–1113. https://doi.org/10.1002/prot.22111
Qu Y, Bolen DW (2002) Efficacy of macromolecular crowding in forcing proteins to fold. Biophys Chem 101–102:155–165. https://doi.org/10.1016/S0301-4622(02)00148-5
Mukherjee S, Waegele MM, Chowdhury P et al (2009) Effect of macromolecular crowding on protein folding dynamics at the secondary structure level. J Mol Biol 393:227–236. https://doi.org/10.1016/j.jmb.2009.08.016
Cheung MS, Klimov D, Thirumalai D (2005) Molecular crowding enhances native state stability and refolding rates of globular proteins. Proc Natl Acad Sci U S A 102:4753–4758. https://doi.org/10.1073/pnas.0409630102
Perham M, Stagg L, Wittung-Stafshede P (2007) Macromolecular crowding increases structural content of folded proteins. FEBS Lett 581:5065–5069. https://doi.org/10.1016/j.febslet.2007.09.049
Zhou BR, Liang Y, Du F et al (2004) Mixed macromolecular crowding accelerates the oxidative refolding of reduced, denatured lysozyme: Implications for protein folding in intracellular environments. J Biol Chem 279:55109–55116. https://doi.org/10.1074/jbc.M409086200
Acknowledgements
The authors are highly thankful for the facilities obtained at Department of Biochemistry, AMU Aligarh.
Author information
Authors and Affiliations
Contributions
Experimental work done and Manuscript written by Gufran Ahmed Siddiqui, Manuscript checked and supervised by Dr Aabgeena Naeem.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declared that there is no conflict of interest.
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Siddiqui, G.A., Naeem, A. Refolding of Hemoglobin Under Macromolecular Confinement: Impersonating In Vivo Volume Exclusion. J Fluoresc 31, 1371–1377 (2021). https://doi.org/10.1007/s10895-021-02751-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10895-021-02751-y