Abstract
Purpose
Chromosome 22q11.2 deletion syndrome is a common inborn error of immunity. The early consequences of thymic hypoplasia are low T cell numbers. Later in life, atopy, autoimmunity, inflammation, and evolving hypogammaglobulinemia can occur and the causes of these features are not understood. This study utilized an unbiased discovery approach to define alterations in histone modifications. Our goal was to identify durable chromatin changes that could influence cell behavior.
Methods
CD4 T cells and CD19 B cells underwent ChIP-seq analysis using antibodies to H3K4me3, H3K27ac, and H4ac. RNA effects were defined in CD4 T cells by RNA-seq. Serum cytokines were examined by Luminex.
Results
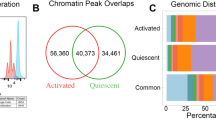
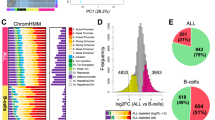
Histone marks of transcriptional activation at CD4 T cell promoters and enhancers were globally increased. The promoter activation signature had elements related to T cell activation and inflammation, concordant with effects seen in the transcriptome. B cells, in contrast, had a minimally altered epigenetic landscape in 22q11.2. Both cell types had an “edge” effect with markedly altered chromatin adjacent to the deletion.
Conclusions
People with 22q11.2 deletion have altered CD4 T cell chromatin and a transcriptome concordant with the changes in the epigenome. These effects support a disease model where qualitative changes to T cells occur in addition to quantitative defects that have been well characterized. This study offers unique insight into qualitative differences in the T cells in 22q11.2 deletion, an aspect that has received limited attention.







Similar content being viewed by others
Data Availability
Data is available upon request.
Code Availability
N/A.
References
Kobrynski LJ, Sullivan KE. Velocardiofacial syndrome, DiGeorge syndrome: the chromosome 22q11.2 deletion syndromes. Lancet. 2007;370(9596):1443–52.
McDonald-McGinn DM, Sullivan KE, Marino B, Philip N, Swillen A, Vorstman JA, et al. 22q11.2 deletion syndrome. Nat Rev Dis Primers. 2015;1:15071.
Piliero LM, Sanford AN, McDonald-McGinn DM, Zackai EH, Sullivan KE. T-cell homeostasis in humans with thymic hypoplasia due to chromosome 22q11.2 deletion syndrome. Blood. 2004;103(3):1020–5.
Dar N, Gothelf D, Korn D, Frisch A, Weizman A, Michaelovsky E, et al. Thymic and bone marrow output in individuals with 22q11.2 deletion syndrome. Pediatr Res. 2015;77(4):579–85.
Ferrando-Martinez S, Lorente R, Gurbindo D, De Jose MI, Leal M, Munoz-Fernandez MA, et al. Low thymic output, peripheral homeostasis deregulation, and hastened regulatory T cells differentiation in children with 22q11.2 deletion syndrome. J Pediatr. 2014;164(4):882–9.
Montin D, Marolda A, Licciardi F, Robasto F, Di Cesare S, Ricotti E, et al. Immunophenotype anomalies predict the development of autoimmune cytopenia in 22q11.2 deletion syndrome. J Allergy Clin Immunol Pract. 2019;7(7):2369–76.
Morsheimer M, Brown Whitehorn TF, Heimall J, Sullivan KE. The immune deficiency of chromosome 22q11.2 deletion syndrome. Am J Med Genet A. 2017;173(9):2366–72.
Giardino G, Radwan N, Koletsi P, Morrogh DM, Adams S, Ip W, et al. Clinical and immunological features in a cohort of patients with partial DiGeorge syndrome followed at a single center. Blood. 2019;133(24):2586–96.
Herwadkar A, Gennery AR, Moran AS, Haeney MR, Arkwright PD. Association between hypoparathyroidism and defective T cell immunity in 22q11.2 deletion syndrome. J Clin Pathol. 2010;63(2):151–5.
Milner JD, Ward JM, Keane-Myers A, Paul WE. Lymphopenic mice reconstituted with limited repertoire T cells develop severe, multiorgan, Th2-associated inflammatory disease. Proc Natl Acad Sci USA. 2007;104(2):576–81.
Campbell JM, Knutsen AP, Becker BA. A 39-year-old father is diagnosed in adulthood as having partial DiGeorge anomaly with a combined T- and B-cell immunodeficiency after diagnosis of the condition in his daughter. Ann Allergy Asthma Immunol. 2008;100(6):620–1.
Pongpruttipan T, Cook JR, Reyes-Mugica M, Spahr JE, Swerdlow SH. Pulmonary extranodal marginal zone lymphoma of mucosa-associated lymphoid tissue associated with granulomatous inflammation in a child with chromosome 22q11.2 deletion syndrome (DiGeorge syndrome). J Pediatr. 2012;161(5):954–8.
Sood AK, Funkhouser W, Handly B, Weston B, Wu EY. Granulomatous-lymphocytic interstitial lung disease in 22q11.2 deletion syndrome: a case report and literature review. Curr Allergy Asthma Rep. 2018;18(3):14.
Tison BE, Nicholas SK, Abramson SL, Hanson IC, Paul ME, Seeborg FO, et al. Autoimmunity in a cohort of 130 pediatric patients with partial DiGeorge syndrome. J Allergy Clin Immunol. 2011;128(5):1115–7 e1–3.
Jawad AF, Prak EL, Boyer J, McDonald-McGinn DM, Zackai E, McDonald K, et al. A prospective study of influenza vaccination and a comparison of immunologic parameters in children and adults with chromosome 22q11.2 deletion syndrome (digeorge syndrome/velocardiofacial syndrome). J Clin Immunol. 2011;31(6):927–35.
Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74.
Zemble R, Luning Prak E, McDonald K, McDonald-McGinn D, Zackai E, Sullivan K. Secondary immunologic consequences in chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome). Clin Immunol. 2010;136(3):409–18.
Derfalvi B, Maurer K, McDonald McGinn DM, Zackai E, Meng W, Luning Prak ET, et al. B cell development in chromosome 22q11.2 deletion syndrome. Clin Immunol. 2016;163:1–9.
Burren OS, Rubio Garcia A, Javierre BM, Rainbow DB, Cairns J, Cooper NJ, et al. Chromosome contacts in activated T cells identify autoimmune disease candidate genes. Genome Biol. 2017;18(1):165.
Shi L, Song L, Maurer K, Dou Y, Patel VR, Su C, et al. IL-1 Transcriptional responses to lipopolysaccharides are regulated by a complex of RNA binding proteins. J Immunol. 2020;204(5):1334–44.
Morou A, Palmer BE, Kaufmann DE. Distinctive features of CD4+ T cell dysfunction in chronic viral infections. Curr Opin HIV AIDS. 2014;9(5):446–51.
Schulze Zur Wiesch J, Ciuffreda D, Lewis-Ximenez L, Kasprowicz V, Nolan BE, Streeck H, et al. Broadly directed virus-specific CD4+ T cell responses are primed during acute hepatitis C infection, but rapidly disappear from human blood with viral persistence. J Exp Med. 2012;209(1):61–75.
Crawford A, Angelosanto JM, Kao C, Doering TA, Odorizzi PM, Barnett BE, et al. Molecular and transcriptional basis of CD4(+) T cell dysfunction during chronic infection. Immunity. 2014;40(2):289–302.
Bektas A, Schurman SH, Gonzalez-Freire M, Dunn CA, Singh AK, Macian F, et al. Age-associated changes in human CD4(+) T cells point to mitochondrial dysfunction consequent to impaired autophagy. Aging (Albany NY). 2019;11(21):9234–63.
Radens CM, Blake D, Jewell P, Barash Y, Lynch KW. Meta-analysis of transcriptomic variation in T-cell populations reveals both variable and consistent signatures of gene expression and splicing. RNA. 2020;26(10):1320–33.
Weinstein JS, Lezon-Geyda K, Maksimova Y, Craft S, Zhang Y, Su M, et al. Global transcriptome analysis and enhancer landscape of human primary T follicular helper and T effector lymphocytes. Blood. 2014;124(25):3719–29.
Javierre BM, Burren OS, Wilder SP, Kreuzhuber R, Hill SM, Sewitz S, et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell. 2016;167(5):1369–84 e19.
Bothma JP, Garcia HG, Ng S, Perry MW, Gregor T, Levine M. Enhancer additivity and non-additivity are determined by enhancer strength in the Drosophila embryo. Elife. 2015;4.
Staple L, Andrews T, McDonald-McGinn D, Zackai E, Sullivan KE. Allergies in patients with chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome) and patients with chronic granulomatous disease. Pediatr Allergy Immunol. 2005;16(3):226–30.
Di Cesare S, Puliafito P, Ariganello P, Marcovecchio GE, Mandolesi M, Capolino R, et al. Autoimmunity and regulatory T cells in 22q11.2 deletion syndrome patients. Pediatr Allergy Immunol. 2015;26(6):591–4.
Klocperk A, Parackova Z, Bloomfield M, Rataj M, Pokorny J, Unger S, et al. Follicular helper T cells in DiGeorge syndrome. Front Immunol. 2018;9:1730.
Nagakubo D, Swann JB, Birmelin S, Boehm T. Autoimmunity associated with chemically induced thymic dysplasia. Int Immunol. 2017;29(8):385–90.
Aresvik DM, Lima K, Overland T, Mollnes TE, Abrahamsen TG. Increased levels of interferon-inducible protein 10 (IP-10) in 22q11.2 deletion syndrome. Scand J Immunol. 2016;83(3):188–94.
Thauland TJ, Pellerin L, Ohgami RS, Bacchetta R, Butte MJ. Case study: mechanism for increased follicular helper T cell development in activated PI3K delta syndrome. Front Immunol. 2019;10:753.
Choi JY, Ho JH, Pasoto SG, Bunin V, Kim ST, Carrasco S, et al. Circulating follicular helper-like T cells in systemic lupus erythematosus: association with disease activity. Arthritis Rheumatol. 2015;67(4):988–99.
Shen W, Ye H, Zhang X, Huo L, Shen J, Zhu L, et al. Elevated expansion of follicular helper T cells in peripheral blood from children with acute measles infection. BMC Immunol. 2020;21(1):49.
Toubi E, Gordon S, Kessel A, Rosner I, Rozenbaum M, Shoenfeld Y, et al. Elevated serum B-Lymphocyte activating factor (BAFF) in chronic hepatitis C virus infection: association with autoimmunity. J Autoimmun. 2006;27(2):134–9.
Moisini I, Davidson A. BAFF: a local and systemic target in autoimmune diseases. Clin Exp Immunol. 2009;158(2):155–63.
Zhang X, Zhang Y, Zhu X, Purmann C, Haney MS, Ward T, et al. Local and global chromatin interactions are altered by large genomic deletions associated with human brain development. Nat Commun. 2018;9(1):5356.
Fulcoli FG, Franzese M, Liu X, Zhang Z, Angelini C, Baldini A. Rebalancing gene haploinsufficiency in vivo by targeting chromatin. Nat Commun. 2016;7:11688.
de la Morena MT, Eitson JL, Dozmorov IM, Belkaya S, Hoover AR, Anguiano E, et al. Signature MicroRNA expression patterns identified in humans with 22q11.2 deletion/DiGeorge syndrome. Clin Immunol. 2013;147(1):11–22.
Sellier C, Hwang VJ, Dandekar R, Durbin-Johnson B, Charlet-Berguerand N, Ander BP, et al. Decreased DGCR8 expression and miRNA dysregulation in individuals with 22q11.2 deletion syndrome. PLoS One. 2014;9(8):e103884.
Stohl W, Hiepe F, Latinis KM, Thomas M, Scheinberg MA, Clarke A, et al. Belimumab reduces autoantibodies, normalizes low complement levels, and reduces select B cell populations in patients with systemic lupus erythematosus. Arthritis Rheum. 2012;64(7):2328–37.
Acknowledgements
We gratefully acknowledge the 22q and You Center at CHOP and all the patients and families.
Funding
KES is supported by the Wallace Chair of Pediatrics.
Author information
Authors and Affiliations
Contributions
Dr. Zhe performed the sequencing analyses.
Dr. Shi made the ChIP-seq libraries and performed data analysis.
Mss. Song, Maurer, and Zhao did cell purification, cell culture, and analyses.
Dr. Zackai participated in experimental design.
Mr. Crowley and Mr. McGinn performed data analyses, clinical data extraction, and sample preparation.
Ms. McGinn and Dr. Sullivan were responsible for the conception, compliance, and execution of the experiments.
Corresponding author
Ethics declarations
Ethics Approval
This study was approved by the IRB at The Children’s Hospital of Philadelphia.
Consent to Participate
Patients consented to the participation in this study.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, Z., Shi, L., Song, L. et al. Chromatin Modifications in 22q11.2 Deletion Syndrome. J Clin Immunol 41, 1853–1864 (2021). https://doi.org/10.1007/s10875-021-01123-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10875-021-01123-2