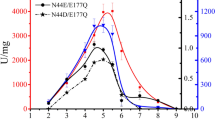
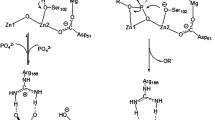
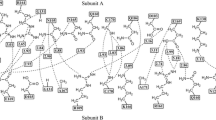
Abstract
NMR-monitored pH titration curves of proteins provide a rich source of structural and electrostatic information. Although relatively straightforward to measure, interpreting pH-dependent chemical shift changes to obtain site-specific acid dissociation constants (pK A values) is challenging. In order to analyze the biphasic titrations exhibited by the side chain 13Cγ nuclei of the nucleophilic Glu78 and general acid/base Glu172 in Bacillus circulans xylanase, we have revisited the formalism for the ionization equilibria of two coupled acidic residues. In general, fitting NMR-monitored pH titration curves for such a system will only yield the two macroscopic pK A values that reflect the combined effects of both deprotonation reactions. However, through the use of mutations complemented with ionic strength-dependent measurements, we are able to extract the four microscopic pK Ai values governing the branched acid/base equilibria of Glu78 and Glu172 in BcX. These data, confirmed through theoretical calculations, help explain the pH-dependent mechanism of this model GH11 xylanase by demonstrating that the kinetically determined pK A values and hence catalytic roles of these two residues result from their electrostatic coupling.






Similar content being viewed by others
References
Alberty RA (2000) Effect of pH on protein-ligand equilibria. J Phys Chem B 104:9929–9934
Andre I, Linse S, Mulder FAA (2007) Residue-specific pK(a) determination of lysine and arginine side chains by indirect 15N and 13C NMR spectroscopy: application to apo calmodulin. J Am Chem Soc 129:15805–15813
Batchelor JG, Feeney J, Roberts GCK (1975) C-13 NMR protonation shifts of amines, carboxylic-acids and amino-acids. J Mag Reson 20:19–38
Blomberg F, Maurer W, Ruterjans H (1977) Nuclear magnetic-resonance investigation of 15N -labeled histidine in aqueous-solution. J Am Chem Soc 99:8149–8159
Buckingham AD (1960) Chemical shifts in the nuclear magnetic resonance spectra of molecules containing polar groups. Can J Chem 38:300–307
Chivers PT, Prehoda KE, Volkman BF, Kim BM, Markley JL, Raines RT (1997) Microscopic pK(a) values of Escherichia coli thioredoxin. Biochemistry 36:14985–14991
Creighton TE (2010) The biophysical chemistry of nucleic acids and proteins. Helvetian Press
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRpipe—a multidimensional spectral processing system based on Unix pipes. J Biomol NMR 6:277–293
Edsall JT, Wyman J (1958) Biophysical chemistry. Academic Press, New York
Farrell D, Miranda ES, Webb H, Georgi N, Crowley PB, McIntosh LP, Nielsen JE (2010) Titration_DB: storage and analysis of NMR-monitored protein pH titration curves. Proteins Struct Func Bioinf 78:843–857
Goddard TD, Kneller DG (2004) Sparky 3. University of California, San Francisco
Hass MA, Jensen MR, Led JJ (2008) Probing electric fields in proteins in solution by NMR spectroscopy. Proteins 72:333–343
Hooft RWW, Sander C, Vriend G (1996) Positioning hydrogen atoms by optimizing hydrogen-bond networks in protein structures. Proteins Struct Func Gen 26:363–376
Joshi MD, Hedberg A, McIntosh LP (1997) Complete measurement of the pK(a) values of the carboxyl and imidazole groups in Bacillus circulans xylanase. Prot Sci 6:2667–2670
Joshi MD, Sidhu G, Nielsen JE, Brayer GD, Withers SG, McIntosh LP (2001) Dissecting the electrostatic interactions and pH-dependent activity of a family 11 glycosidase. Biochemistry 40:10115–10139
Kay L (1993) Pulsed-field gradient-enhanced three-dimensional NMR experiment for correlating 13Cα/β, 13C′, and 1Hα chemical shifts in uniformly 13C labelled proteins dissolved in H2O. J Am Chem Soc 115:2055–2057
Klingen AR, Bombarda E, Ullmann GM (2006) Theoretical investigation of the behavior of titratable groups in proteins. Photochem Photobiol Sci 5:588–596
Lindman S, Linse S, Mulder FAA, Andre I (2006) Electrostatic contributions to residue-specific protonation equilibria and proton binding capacitance for a small protein. Biochemistry 45:13993–14002
Markley JL (1975) Observation of histidine residues in proteins by means of nuclear magnetic-resonance spectroscopy. Acc Chem Res 8:70–80
McIntosh LP, Hand G, Johnson PE, Joshi MD, Korner M, Plesniak LA, Ziser L, Wakarchuk WW, Withers SG (1996) The pK(a) of the general acid/base carboxyl group of a glycosidase cycles during catalysis: a 13C-NMR study of Bacillus circuluns xylanase. Biochemistry 35:9958–9966
Nielsen JE (2009) Analyzing enzymatic pH activity profiles and protein titration curves using structure-based pKa calculations and titration curve fitting. Methods Enzymol 454:233–258
Nielsen JE, Andersen KV, Honig B, Hooft RWW, Klebe G, Vriend G, Wade RC (1999) Improving macromolecular electrostatics calculations. Prot Eng 12:657–662
Oda Y, Yamazaki T, Nagayama K, Kanaya S, Kuroda Y, Nakamura H (1994) Individual ionization-constants of all the carboxyl groups in ribonuclease Hi from Escherichia-coli determined by NMR. Biochemistry 33:5275–5284
Onufriev A, Case DA, Ullmann GM (2001) A novel view of pH titration in biomolecules. Biochemistry 40:3413–3419
Pace CN, Grimsley GR, Scholtz JM (2009) Protein ionizable groups: pK values and their contribution to protein stability and solubility. J Biol Chem 284:13285–13289
Pelton JG, Torchia DA, Meadow ND, Roseman S (1993) Tautomeric states of the active-site histidines of phosphorylated and unphosphorylated Iii(Glc), a signal-transducing protein from Escherichia coli, using 2-dimensional heteronuclear NMR techniques. Prot Sci 2:543–558
Plesniak LA, Wakarchuk WW, McIntosh LP (1996) Secondary structure and NMR assignments of Bacillus circulans xylanase. Prot Sci 5:1118–1135
Quirt AR, Lyerla JR, Peat IR, Cohen JS, Reynolds WF, Freedman MH (1974) 13C Nuclear magnetic-resonance titration shifts in amino-acids. J Am Chem Soc 96:570–571
Rabenstein DL, Sayer TL (1976a) 13C Chemical-shift parameters for amines, carboxylic-acids, and amino-acids. J Mag Reson 24:27–39
Rabenstein DL, Sayer TL (1976b) Determination of microscopic acid dissociation-constants by nuclear magnetic-resonance spectrometry. Anal Chem 48:1141–1145
Rouxfromy M (1982) On the hill plot of NMR data for titration of protein residues. Biophys Struct Mech 8:289–306
Sattler M, Schleucher J, Griesinger C (1999) Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Mag Reson Spec 34:93–158
Shrager RI, Sachs DH, Schechte A, Cohen JS, Heller SR (1972) Nuclear magnetic-resonance titration curves of histidine ring protons. 2. Mathematical models for interacting groups in nuclear magnetic-resonance titration curves. Biochemistry 11:541–547
Sondergaard CR, McIntosh LP, Pollastri G, Nielsen JE (2008) Determination of electrostatic interaction energies and protonation state populations in enzyme active sites. J Mol Biol 376:269–287
Sudmeier JL, Reilley CN (1964) Nuclear magnetic resonance studies of protonation of polyamine + aminocarboxylate compounds in aqueous solution. Anal Chem 36:1698
Surprenant HL, Sarneski JE, Key RR, Byrd JT, Reilley CN (1980) 13C NMR-studies of amino-acids—chemical-shifts, protonation shifts, microscopic protonation behavior. J Mag Reson 40:231–243
Szakacs Z, Kraszni M, Noszal B (2004) Determination of microscopic acid-base parameters from NMR-pH titrations. Anal Bioanal Chem 378:1428–1448
Tomlinson JH, Green VL, Baker PJ, Williamson MP (2010) Structural origins of pH-dependent chemical shifts in the B1 domain of protein G. Proteins Struct Func Bioinf 78:3000–3016
Ullmann GM (2003) Relations between protonation constants and titration curves in polyprotic acids: a critical view. J Phys Chem B 107:1263–1271
Wang ZX (1995) An exact mathematical expression for describing competitive-binding of 2 different ligands to a protein molecule. FEBS Lett 360:111–114
Webb H, Tynan-Connolly BM, Lee GM, Farrell D, O’Meara F, Sondergaard CR, Teilum K, Hewage C, McIntosh LP, Nielsen JE (2011) Remeasuring HEWL pK(a) values by NMR spectroscopy: methods, analysis, accuracy, and implications for theoretical pK(a), calculations. Proteins Struct Func Bioinf 79:685–702
Yang AS, Gunner MR, Sampogna R, Sharp K, Honig B (1993) On the calculation of pK(a)s in proteins. Proteins Struc Func Gene 15:252–265
Acknowledgments
We are grateful Lewis Kay for friendship, expert advice, and countless pulse sequences, without which none of our work would be possible. Yom Huledet Same’ach! We also thank Rick Dahlquist and Steve Withers for insightful comments and Philip Johnson for early experimental help. This research was funded by the Natural Sciences and Engineering Research Council of Canada (NSERC; to LPM). Instrument support was provided by the Canadian Institutes for Health Research (CIHR), the Canadian Foundation for Innovation (CFI), the British Columbia Knowledge Development Fund (BCKDF), the UBC Blusson Fund, and the Michael Smith Foundation for Health Research (MSFHR).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
McIntosh, L.P., Naito, D., Baturin, S.J. et al. Dissecting electrostatic interactions in Bacillus circulans xylanase through NMR-monitored pH titrations. J Biomol NMR 51, 5 (2011). https://doi.org/10.1007/s10858-011-9537-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10858-011-9537-x