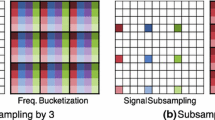
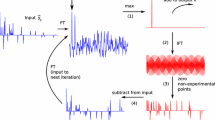
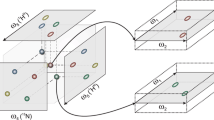
Abstract
Spectra obtained by application of multidimensional Fourier Transformation (MFT) to sparsely sampled nD NMR signals are usually corrupted due to missing data. In the present paper this phenomenon is investigated on simulations and experiments. An effective iterative algorithm for artifact suppression for sparse on-grid NMR data sets is discussed in detail. It includes automated peak recognition based on statistical methods. The results enable one to study NMR spectra of high dynamic range of peak intensities preserving benefits of random sampling, namely the superior resolution in indirectly measured dimensions. Experimental examples include 3D 15N- and 13C-edited NOESY-HSQC spectra of human ubiquitin.










Similar content being viewed by others
References
Armstrong GS, Mandelshtam VA, Shaka AJ, Bendiak B (2005) Rapid high-resolution four-dimensional NMR spectroscopy using the filter diagonalization method and its advantages for detailed structural elucidation of oligosaccharides. J Magn Reson 173:160–168
Barna JCJ, Tan SM, Laue ED (1988) Use of CLEAN in conjunction with selective data sampling for 2D NMR experiments. J Magn Reson 78:327–332
Blackford LS, Demmel J, Dongarra J, Duff I, Hammarling S, Henry G, Heroux M, Kaufman L, Lumsdaine A, Petitet A, Pozo R, Remington K, Whaley RC (2002) An updated set of basic linear algebra subprograms (BLAS). ACM Trans Math Soft 28:135–151
Bodenhausen G, Ernst RR (1981) The accordion experiment, a simple approach to three-dimensional NMR spectroscopy. J Magn Reson 45:367–373
Bracewell RN (2000) The Fourier transform and its applications. McGraw-Hill Higher Education, New York
Coggins BE, Zhou P (2007) Sampling of the NMR time domain along concentric rings. J Magn Reson 184:207–221
Coggins BE, Zhou P (2008) High resolution 4-D spectroscopy with sparse concentric shell sampling and FFT-CLEAN. J Biomol NMR 42:225–239
Ding K, Gronenborn AM (2002) Novel 2D triple-resonance NMR experiments for sequential resonance assignments of proteins. J Magn Reson 156:262–268
Eghbalnia HR, Bahrami A, Tonelli M, Hallenga K, Markley JL (2005) High-resolution iterative frequency identification for NMR as a general strategy for multidimensional data collection. J Am Chem Soc 127:12528–12536
Frigo M, Johnson SG (2005) The design and implementation of FFTW3. Proc IEEE 93:216–231
Frydman L, Scherf T, Lupulescu A (2002) The acquisition of multidimensional NMR spectra within a single scan. Proc Natl Acad Sci 99:15662–15858
Goddart TD, Kneller DG (1989–2008) SPARKY 3. University of California, San Francisco
Hiller S, Fiorito F, Wüthrich, Wider G (2005) Automated projection spectroscopy (APSY). P Natl Acad Sci USA 102:10876–10881
Hoch JC, Stern AS (1996) NMR data processing. Wiley, New York
Högbom J (1974) Aperture synthesis with a non-regular distribution of interferometer baselines. Astron Astrophys Suppl 15:417–426
Hyberts SG, Frueh DP, Arthanari H, Wagner G (2009) FM reconstruction of non-uniformly sampled protein NMR data at higher dimensions and optimization by distillation. J Biomol NMR 45:283–294
Kazimierczuk K, Zawadzka A, Koźmiński W, Zhukov I (2006) Random sampling of evolution time space and Fourier transform processing. J Biomol NMR 36:157–168
Kazimierczuk K, Zawadzka A, Koźmiński W, Zhukov I (2007) Lineshapes and artifacts in multidimensional Fourier transform of arbitrary sampled NMR data sets. J Magn Reson 188:344–356
Kazimierczuk K, Zawadzka A, Koźmiński W (2008a) Optimization of random time domain sampling in multidimensional NMR. J Magn Reson 192:123–130
Kazimierczuk K, Zawadzka A, Koźmiński W, Zhukov I (2008b) Determination of spin-spin couplings from ultrahigh resolution 3D NMR spectra obtained by optimized random sampling and multidimensional Fourier transformation. J Am Chem Soc 130:5404–5405
Kazimierczuk K, Zawadzka A, Koźmiński W (2009) Narrow peaks and high dimensionalities: exploiting the advantages of random sampling. J Magn Reson 197:219–228
Kim S, Szyperski T (2003) GFT NMR, a new approach to rapidly obtain precise high-dimensional NMR spectral information. J Am Chem Soc 125:1385–1393
Koźmiński W, Zhukov I (2003) Multiple quadrature detection in reduced dimensionality experiments. J Biomol NMR 26:157–166
Kupče E, Freeman R (2003a) Projection-reconstruction of three-dimensional NMR spectra. J Am Chem Soc 125:13958–13959
Kupče E, Freeman R (2003b) Reconstruction of the three-dimensional NMR spectrum of a protein from a set of plane projection. J Biomol NMR 27:383–387
Kupče E, Freeman R (2005) Fast multidimensional NMR: radial sampling of evolution space. J Magn Reson 173:317–321
Luan T, Jaravine V, Yee A, Arrowsmith CH, Orekhov VY (2005) Optimization of resolution and sensitivity of 4D NOESY using multidimensional decomposition. J Biomol NMR 33:1–14
Malmodin D, Billeter M (2005a) Multiway decomposition of NMR spectra with coupled evolution periods. J Am Chem Soc 127:13486–13487
Malmodin D, Billeter M (2005b) Signal identification in NMR spectra with coupled evolution periods. J Magn Reson 176:47–53
Mandelshtam VA, Taylor HS, Shaka AJ (1998) Application of the filter diagonalization method to one- and two-dimensional NMR spectra. J Magn Reson 133:304–312
Marion D (2005) Fast acquisition of NMR spectra using fourier transform of non-equispaced data. J Biomol NMR 32:141–150
Marion D (2006) Processing of ND NMR spectra sampled in polar coordinates: a simple fourier transform instead of reconstruction. J Biomol NMR 36:45–54
Matsuki Y, Eddy MT, Herzfeld J (2009) Spectroscopy by integration of frequency and time domain information for fast acquisition of high-resolution dark spectra. J Am Chem Soc 131:4648–4656
Mobli M, Stern A, Hoch JC (2006) Spectral reconstruction methods in fast NMR: reduced dimensionality, random sampling and maximum entropy. J Magn Reson 182:96–105
Mobli M, Maciejewski MW, Gryk MR, Hoch JC (2007) Automatic maximum entropy spectral reconstruction in NMR. J Biomol NMR 39:133–139
Orekhov VY, Ibraghimov I, Billeter M (2003) Optimizing resolution in multidimensional NMR by three-way decomposition. J Biomol NMR 27:165–173
Press WH, Flannery BP, Teukolsky SA, Vetterling WT (2007) Numerical recipes in C, 3rd edn. Cambridge University Press, Cambridge
Rovnyak D, Frueh DP, Sastry M, Sun ZYJ, Stern AS, Hoch JC, Wagner G (2004) Accelerated acquisition of high resolution triple-resonance spectra using non-uniform sampling and maximum entropy reconstruction. J Magn Reson 170:15–21
Schanda P, Melckebeke HV, Brutscher B (2006) Speeding up three-dimensional protein NMR experiments to a few minutes. J Am Chem Soc 128:9042–9043
Szyperski T, Yeh DC, Sukumaran DK, Moseley HNB, Montelione GT (2002) Reduced-dimensionality NMR spectroscopy for high-throughput protein resonance assignment. P Natl Acad Sci USA 99:8009–8014
Tugarinov V, Kay LE, Ibraghimov I, Orekhov VY (2005) Highresolution four-dimensional H-1-C-13 NOE spectroscopy using methyl-TROSY, sparse data acquisition, and multidimensional decomposition. J Am Chem Soc 127:2767–2775
Zawadzka-Kazimierczuk A, Kazimierczuk K, Koźmiński W (2010) A set of 4D NMR experiments of enhanced resolution for easy resonance assignment in proteins. J Magn Reson 202:109–116
Acknowledgments
Special thanks are addressed at Maxim Mayzel from Swedish NMR Centre for his help in using MDD package, and at prof. J.C. Hoch from University of Connecticut Health Center for providing access to Rowland NMR Toolkit v.3. This work was supported by grant number: N301 07131/2159, founded by Ministry of Science and Higher Education in years 2006-2009. Research cofinanced by the European Social Fund and State funds under the Integrated Regional Operational Programme, Measure 2.6 “Regional Innovation Strategies and transfer of knowledge”, Mazovian Voivodship project “Mazovian Ph.D. Scholarship”.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Stanek, J., Koźmiński, W. Iterative algorithm of discrete Fourier transform for processing randomly sampled NMR data sets. J Biomol NMR 47, 65–77 (2010). https://doi.org/10.1007/s10858-010-9411-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-010-9411-2