Abstract
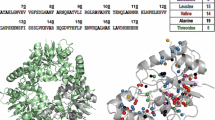
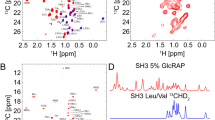
Reductive methylation of lysine residues in proteins offers a way to introduce 13C methyl groups into otherwise unlabeled molecules. The 13C methyl groups on lysines possess favorable relaxation properties that allow highly sensitive NMR signal detection. One of the major limitations in the use of reductive methylation in NMR is the signal overlap of 13C methyl groups in NMR spectra. Here we show that the uniform influence of the solvent on chemical shifts of exposed lysine methyl groups could be overcome by adjusting the pH of the buffering solution closer to the pKa of lysine side chains. Under these conditions, due to variable pKa values of individual lysine side chains in the protein of interest different levels of lysine protonation are observed. These differences are reflected in the chemical shift differences of methyl groups in reductively methylated lysines. We show that this approach is successful in four different proteins including Ca2+-bound Calmodulin, Lysozyme, Ca2+-bound Troponin C, and Glutathione S-Transferase. In all cases significant improvement in NMR spectral resolution of methyl signals in reductively methylated proteins was obtained. The increased spectral resolution helps with more precise characterization of protein structural rearrangements caused by ligand binding as shown by studying binding of Calmodulin antagonist trifluoperazine to Calmodulin. Thus, this approach may be used to increase resolution in NMR spectra of 13C methyl groups on lysine residues in reductively methylated proteins that enhances the accuracy of protein structural assessment.






Similar content being viewed by others
Abbreviations
- CaM:
-
Calmodulin
- HSQC:
-
Heteronuclear Single Quantum Coherence
- GST:
-
Glutathione S-Transferase
- TFP:
-
Trifluoperazine
References
Abraham SJ, Hoheisel S, Gaponenko V (2008) Detection of protein-ligand interactions by NMR using reductive methylation of lysine residues. J Biomol NMR 42:143–148
Andre I, Linse S, Mulder FA (2007) Residue-specific pKa determination of lysine and arginine side chains by indirect 15 N and 13C NMR spectroscopy: application to apo calmodulin. J Am Chem Soc 129:15805–15813
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Dixon MM, Brennan RG, Matthews BW (1991) Structure of gamma-chymotrypsin in the range pH 2.0 to pH 10.5 suggests that gamma-chymotrypsin is a covalent acyl-enzyme adduct at low pH. Int J Biol Macromol 13:89–96
Fujita T, Ralston GB, Morris MB (1998) Biophysical properties of human erythrocyte spectrin at alkaline pH: implications for spectrin structure, function, and association. Biochemistry 37:264–271
Ganzhorn AJ, Lepage P, Pelton PD, Strasser F, Vincendon P, Rondeau JM (1996) The contribution of lysine-36 to catalysis by human myo-inositol monophosphatase. Biochemistry 35:10957–10966
Gao G, Prasad R, Lodwig SN, Unkefer CJ, Beard WA, Wilson SH, London RE (2006) Determination of lysine pK values using [5–13C]lysine: application to the lyase domain of DNA Pol beta. J Am Chem Soc 128:8104–8105
Gerken TA, Jentoft JE, Jentoft N, Dearborn DG (1982) Intramolecular interactions of amino groups in 13C reductively methylated hen egg-white lysozyme. J Biol Chem 257:2894–2900
Gopalakrishna R, Anderson WB (1982) Ca2+-induced hydrophobic site on calmodulin: application for purification of calmodulin by phenyl-Sepharose affinity chromatography. Biochem Biophys Res Commun 104:830–836
Harmat V, Bocskei Z, Naray-Szabo G, Bata I, Csutor AS, Hermecz I, Aranyi P, Szabo B, Liliom K, Vertessy BG, Ovadi J (2000) A new potent calmodulin antagonist with arylalkylamine structure: crystallographic, spectroscopic and functional studies. J Mol Biol 297:747–755
Harries WE, Akhavan D, Miercke LJ, Khademi S, Stroud RM (2004) The channel architecture of aquaporin 0 at a 2.2-A resolution. Proc Natl Acad Sci U S A 101:14045–14050
Iwahara J, Jung YS, Clore GM (2007) Heteronuclear NMR spectroscopy for lysine NH(3) groups in proteins: unique effect of water exchange on (15)N transverse relaxation. J Am Chem Soc 129:2971–2980
Kesvatera T, Jonsson B, Thulin E, Linse S (1996) Measurement and modelling of sequence-specific pKa values of lysine residues in calbindin D9k. J Mol Biol 259:828–839
Kobayashi T, Solaro RJ (2006) Increased Ca2+ affinity of cardiac thin filaments reconstituted with cardiomyopathy-related mutant cardiac Troponin I. J Biol Chem 281:13471–13477
Kurinov IV, Mao C, Irvin JD, Uckun FM (2000) X-ray crystallographic analysis of pokeweed antiviral protein-II after reductive methylation of lysine residues. Biochem Biophys Res Commun 275:549–552
Macnaughtan MA, Kane AM, Prestegard JH (2005) Mass spectrometry assisted assignment of NMR resonances in reductively 13C-methylated proteins. J Am Chem Soc 127:17626–17627
Means GE, Feeney RE (1968) Reductive alkylation of amino groups in proteins. Biochemistry 7:2192–2201
Rayment I (1997) Reductive alkylation of lysine residues to alter crystallization properties of proteins. Methods Enzymol 276:171–179
Rypniewski WR, Holden HM, Rayment I (1993) Structural consequences of reductive methylation of lysine residues in hen egg white lysozyme: an X-ray analysis at 1.8-A resolution. Biochemistry 32:9851–9858
Takayama Y, Castaneda CA, Chimenti M, Garcia-Moreno B, Iwahara J (2008) Direct evidence for deprotonation of a lysine side chain buried in the hydrophobic core of a protein. J Am Chem Soc 130:6714–6715
Venugopal MG, Wolfersberger MG, Wallace BA (1992) Effects of pH on conformational properties related to the toxicity of Bacillus thuringiensis delta-endotoxin. Biochim Biophys Acta 1159:185–192
Walter TS, Meier C, Assenberg R, Au KF, Ren J, Verma A, Nettleship JE, Owens RJ, Stuart DI, Grimes JM (2006) Lysine methylation as a routine rescue strategy for protein crystallization. Structure 14:1617–1622
Zhang M, Vogel HJ (1993) Determination of the side chain pKa values of the lysine residues in calmodulin. J Biol Chem 268:22420–22428
Acknowledgements
We are grateful to Dr. Arnon Lavie and Ms. Ming F. Lye for providing purified GST for our experiments. The research was supported by the American Cancer Society, Illinois Division Grant ACS 08-14 to Vadim Gaponenko, NIH Grant R01HL082923 to Tomoyoshi Kobayashi, and P01HL62426 to R. John Solaro. NMR spectra were recorded using the NMR facility at the University of Illinois at Chicago funded by the NIH Grant P41 GM68944 to Peter GW Gettins.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Abraham, S.J., Kobayashi, T., John Solaro, R. et al. Differences in lysine pKa values may be used to improve NMR signal dispersion in reductively methylated proteins. J Biomol NMR 43, 239–246 (2009). https://doi.org/10.1007/s10858-009-9306-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-009-9306-2