Abstract
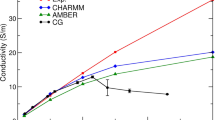
In this paper, we present a full three-dimensional simulation of the ompF porin channel using BioMOCA, a self-consistent particle-based ion channel simulation tool, based on the Boltzmann Transport Monte Carlo methodology widely used to simulate conduction in the solid-state device. Significant computational speed-up over atomistic Molecular Dynamics simulations is achieved by treating protein, membrane and water as continuum dielectric background media and computing only the trajectories of mobile ions in solution. A realistic channel structure with permanent fixed charges is mapped onto a finite mesh using the Cloud-in-Cell scheme. Electrostatic forces, computed by solving Poisson equation at regular intervals, are added to a pair-wise ion-ion interaction, which is necessary to prevent the unphysical coalescence of finite-sized ions. The interaction between ions and water is modeled as a random scattering process that thermalizes the ion. Using this tool we computed the complete current-voltage characteristic of the porin channel in approximately one week using ten IBM p690 processors. We also present steady-state ion channel occupancies and compare them with results obtained from recent drift-diffusion based simulations.
Similar content being viewed by others
References
B. Hille, Ion Channels of Excitable Membranes (Sinauer Associates Inc., Sunderland, 2001).
D. Aidley and P.R. Stanfield, Ion Channels: Molecules in Action (Cambridge University Press, Cambridge, 1996).
D.P. Tieleman et al., “Simulation approaches to ion channel structure-function relationships,” Quarterly Reviews of Biophysics, 34, 473 (2001).
J.M. Haile, Molecular Dynamics Simulation: Elementary Methods (John Wiley and Sons, New York, 1997).
S. Selberherr, Analysis and Simulation of Semiconductor Devices (Springer-Verlag, Vienna, 1984).
T. van der Straaten et al., “BioMOCA: A Transport Monte Carlo Model for Ion Channels,” Journal of Computational Electronics, 2, 231 (2003).
T. van der Straaten et al., “BioMOCA: A Boltzmann Transport Monte Carlo Model for Ion Channel Simulation,” Molecular Simulation, in press.
S.W. Cowan, “Bacterial Porins-Lessons from 3 High-resolution Structures,” Current Opinion in Structural Biology, 3, 501 (1993).
R.W. Hockney and J.W. Eastwood, Computer Simulation Using Particles (McGraw-Hill, New York, 2001).
D. Sitkoff et al., “Accurate Calculation of Hydration Free Energies Using Macroscopic Solvent Models,” Journal of Physical Chemistry, 98, 1978 (1994).
T. van der Straaten et al., “Simulating Ion Permeation Through the ompF Porin Ion Channel Using Three-Dimensional Drift-Diffusion Theory,” Journal of Computational Electronics, 2, 29 (2003).
W. Nonner et al., “Progress and Prospects in Permeation,” Journal of General Physiology, 113, 773 (1999).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Lee, KI., Park, Y.J., Der Straaten, T.V. et al. Simulation of Ion Conduction in the ompF Porin Channel Using BioMOCA. J Comput Electron 4, 157–160 (2005). https://doi.org/10.1007/s10825-005-7129-2
Issue Date:
DOI: https://doi.org/10.1007/s10825-005-7129-2