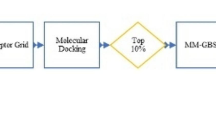
Abstract
The Wnt/β-catenin signaling pathway plays a significant role in the control of osteoblastogenesis and bone formation. CXXC finger protein 5 (CXXC5) has been recently identified as a negative feedback regulator of osteoblast differentiation through a specific interaction with Dishevelled (Dvl) protein. It was reported that targeting the Dvl–CXXC5 interaction could be a novel anabolic therapeutic target for osteoporosis. In this study, complex structure of Dvl PDZ domain and CXXC5 peptide was simulated with molecular dynamics (MD). Based on the structural analysis of binding modes of MD-simulated Dvl PDZ domain with CXXC5 peptide and crystal Dvl PDZ domain with synthetic peptide–ligands, we generated two different pharmacophore models and applied pharmacophore-based virtual screening to discover potent inhibitors of the Dvl–CXXC5 interaction for the anabolic therapy of osteoporosis. Analysis of 16 compounds selected by means of a virtual screening protocol yielded four compounds that effectively disrupted the Dvl–CXXC5 interaction in the fluorescence polarization assay. Potential compounds were validated by fluorescence spectroscopy and nuclear magnetic resonance. We successfully identified a highly potent inhibitor, BMD4722, which directly binds to the Dvl PDZ domain and disrupts the Dvl–CXXC5 interaction. Overall, CXXC5–Dvl PDZ domain complex based pharmacophore combined with various traditional and simple computational methods is a promising approach for the development of modulators targeting the Dvl–CXXC5 interaction, and the potent inhibitor BMD4722 could serve as a starting point to discover or design more potent and specific the Dvl–CXXC5 interaction disruptors.









Similar content being viewed by others
References
Long F (2012) Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol 13(1):27–38
Dees C, Distler JH (2013) Canonical Wnt signalling as a key regulator of fibrogenesis: implications for targeted therapies?. Exp Dermatol 22(11):710–713. https://doi.org/10.1111/exd.12255
Schinner S (2009) Wnt-signalling and the metabolic syndrome. Horm Metab Res 41(2):159–163. https://doi.org/10.1055/s-0028-1119408
Berwick DC, Harvey K (2012) The importance of Wnt signalling for neurodegeneration in Parkinson’s disease. Biochem Soc Trans 40(5):1123–1128. https://doi.org/10.1042/bst20120122
Inestrosa NC, Montecinos-Oliva C, Fuenzalida M (2012) Wnt signaling: role in Alzheimer disease and schizophrenia. J Neuroimmune Pharmacol 7(4):788–807. https://doi.org/10.1007/s11481-012-9417-5
Rachner TD, Khosla S, Hofbauer LC (2011) Osteoporosis: now and the future. Lancet 377(9773):1276–1287. https://doi.org/10.1016/s0140-6736(10)62349-5
Regard JB, Zhong Z, Williams BO, Yang Y (2012) Wnt signaling in bone development and disease: making stronger bone with Wnts. Cold Spring Harb Perspect Biol. https://doi.org/10.1101/cshperspect.a007997
Ke HZ, Richards WG, Li X, Ominsky MS (2012) Sclerostin and Dickkopf-1 as therapeutic targets in bone diseases. Endocr Rev 33(5):747–783. https://doi.org/10.1210/er.2011-1060
Kim HY, Yoon JY, Yun JH, Cho KW, Lee SH, Rhee YM, Jung HS, Lim HJ, Lee H, Choi J, Heo JN, Lee W, No KT, Min D, Choi KY (2015) CXXC5 is a negative-feedback regulator of the Wnt/[beta]-catenin pathway involved in osteoblast differentiation. Cell Death Differ 22(6):912–920. https://doi.org/10.1038/cdd.2014.238
Andersson T, Södersten E, Duckworth JK, Cascante A, Fritz N, Sacchetti P, Cervenka I, Bryja V, Hermanson O (2009) CXXC5 Is a Novel BMP4-regulated modulator of Wnt signaling in neural stem cells. J Biol Chem 284(6):3672–3681. https://doi.org/10.1074/jbc.M808119200
Kim MS, Yoon SK, Bollig F, Kitagaki J, Hur W, Whye NJ, Wu YP, Rivera MN, Park JY, Kim HS, Malik K, Bell DW, Englert C, Perantoni AO, Lee SB (2010) A novel Wilms tumor 1 (WT1) target gene negatively regulates the WNT signaling pathway. J Biol Chem 285(19):14585–14593. https://doi.org/10.1074/jbc.M109.094334
Knappskog S, Myklebust LM, Busch C, Aloysius T, Varhaug JE, Lonning PE, Lillehaug JR, Pendino F (2011) RINF (CXXC5) is overexpressed in solid tumors and is an unfavorable prognostic factor in breast cancer. Ann Oncol 22(10):2208–2215. https://doi.org/10.1093/annonc/mdq737
Shan J, Shi DL, Wang J, Zheng J (2005) Identification of a specific inhibitor of the dishevelled PDZ domain. Biochemistry 44(47):15495–15503. https://doi.org/10.1021/bi0512602
Grandy D, Shan J, Zhang X, Rao S, Akunuru S, Li H, Zhang Y, Alpatov I, Zhang XA, Lang RA, Shi DL, Zheng JJ (2009) Discovery and characterization of a small molecule inhibitor of the PDZ domain of dishevelled. J Biol Chem 284(24):16256–16263. https://doi.org/10.1074/jbc.M109.009647
Shan J, Zheng JJ (2009) Optimizing Dvl PDZ domain inhibitor by exploring chemical space. J Comput-Aided Mol Des 23(1):37–47. https://doi.org/10.1007/s10822-008-9236-1
Choi J, Ma S, Kim H-Y, Yun J-H, Heo J-N, Lee W, Choi K-Y, No KT (2016) Identification of small-molecule compounds targeting the dishevelled PDZ domain by virtual screening and binding studies. Bioorg Med Chem 24(15):3259–3266. https://doi.org/10.1016/j.bmc.2016.03.026
Fujii N, You L, Xu Z, Uematsu K, Shan J, He B, Mikami I, Edmondson LR, Neale G, Zheng J, Guy RK, Jablons DM (2007) An antagonist of dishevelled protein-protein interaction suppresses β-catenin–dependent tumor cell growth. Can Res 67(2):573–579. https://doi.org/10.1158/0008-5472.can-06-2726
Kim HY, Choi S, Yoon JH, Lim HJ, Lee H, Choi J, Ro EJ, Heo JN, Lee W, No KT, Choi KY (2016) Small molecule inhibitors of the Dishevelled-CXXC5 interaction are new drug candidates for bone anabolic osteoporosis therapy. EMBO Mol Med. https://doi.org/10.15252/emmm.201505714
Shivakumar D, Williams J, Wu Y, Damm W, Shelley J, Sherman W (2010) Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J Chem Theory Comput 6(5):1509–1519. https://doi.org/10.1021/ct900587b
Guo Z, Mohanty U, Noehre J, Sawyer TK, Sherman W, Krilov G (2010) Probing the α-helical structural stability of stapled p53 peptides: molecular dynamics simulations and analysis. Chem Biol Drug Des 75(4):348–359. https://doi.org/10.1111/j.1747-0285.2010.00951.x
Bowers KJ, Chow E, Xu H, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossvary I, Moraes MA, Sacerdoti FD, Salmon JK, Shan Y, Shaw DE (2006) Scalable algorithms for molecular dynamics simulations on commodity clusters. In: Proceedings of the ACM/IEEE Conference on Supercomputing (SC06), Tampa, Florida, November 11–17
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79(2):926–935. https://doi.org/10.1063/1.445869
Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) A smooth particle mesh Ewald method. J Chem Phys 103(19):8577–8593. https://doi.org/10.1063/1.470117
Hoover WG (1985) Canonical dynamics: equilibrium phase-space distributions. Phys Rev A 31(3):1695–1697
Martyna GJ, Tobias DJ, Klein ML (1994) Constant pressure molecular dynamics algorithms. J Chem Phys 101(5):4177–4189. https://doi.org/10.1063/1.467468
Humphreys DD, Friesner RA, Berne BJ (1994) A multiple-time-step molecular dynamics algorithm for macromolecules. J Phys Chem 98(27):6885–6892. https://doi.org/10.1021/j100078a035
Schrodinger LLC (2010) The PyMOL molecular graphics system, version 1.3r1
Lee HJ, Wang NX, Shi DL, Zheng JJ (2009) Sulindac inhibits canonical Wnt signaling by blocking the PDZ domain of the protein Dishevelled. Angew Chem Int Ed 48(35):6448–6452. https://doi.org/10.1002/anie.200902981
Acknowledgements
This work was supported by the Ministry of Knowledge Economy through Korea Research Institute of Chemical Technology (SI-1205, SI-1304, SI-1404), and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2016R1A6A3A04010213).
Author information
Authors and Affiliations
Corresponding author
Additional information
Songling Ma and Jiwon Choi are co-first authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ma, S., Choi, J., Jin, X. et al. Discovery of a small-molecule inhibitor of Dvl–CXXC5 interaction by computational approaches. J Comput Aided Mol Des 32, 643–655 (2018). https://doi.org/10.1007/s10822-018-0118-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-018-0118-x