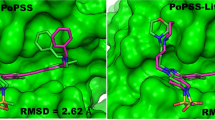
Abstract
Evaluation of ligand three-dimensional (3D) shape similarity is one of the commonly used approaches to identify ligands similar to one or more known active compounds from a library of small molecules. Apart from using ligand shape similarity as a virtual screening tool, its role in pose prediction and pose scoring has also been reported. We have recently developed a method that utilizes ligand 3D shape similarity with known crystallographic ligands to predict binding poses of query ligands. Here, we report the prospective evaluation of our pose prediction method through the participation in drug design data resource (D3R) Grand Challenge 2015. Our pose prediction method was used to predict binding poses of heat shock protein 90 (HSP90) and mitogen activated protein kinase kinase kinase kinase (MAP4K4) ligands and it was able to predict the pose within 2 Å root mean square deviation (RMSD) either as the top pose or among the best of five poses in a majority of cases. Specifically for HSP90 protein, a median RMSD of 0.73 and 0.68 Å was obtained for the top and the best of five predictions respectively. For MAP4K4 target, although the median RMSD for our top prediction was only 2.87 Å but the median RMSD of 1.67 Å for the best of five predictions was well within the limit for successful prediction. Furthermore, the performance of our pose prediction method for HSP90 and MAP4K4 ligands was always among the top five groups. Particularly, for MAP4K4 protein our pose prediction method was ranked number one both in terms of mean and median RMSD when the best of five predictions were considered. Overall, our D3R Grand Challenge 2015 results demonstrated that ligand 3D shape similarity with the crystal ligand is sufficient to predict binding poses of new ligands with acceptable accuracy.
Graphical Abstract






Similar content being viewed by others
References
Tanrikulu Y, Krüger B, Proschak E (2013) The holistic integration of virtual screening in drug discovery. Drug Discov Today 18:358–364
Walters WP, Stahl MT, Murcko MA (1998) Virtual screening—an overview. Drug Discov Today 3:160–178
Kumar A, Zhang KYJ (2015) Hierarchical virtual screening approaches in small molecule drug discovery. Methods 71:26–37
Lavecchia A, Di Giovanni C (2013) Virtual screening strategies in drug discovery: a critical review. Curr Med Chem 20:2839–2860
Muegge I (2008) Synergies of virtual screening approaches. Mini Rev Med Chem 8:927–933
Muegge I, Oloff S (2006) Advances in virtual screening. Drug Discov Today Technol 3:405–411
Drwal MN, Griffith R (2013) Combination of ligand- and structure-based methods in virtual screening. Drug Discov Today Technol 10:e395–e401
Sliwoski G, Kothiwale S, Meiler J, Lowe EW (2014) Computational methods in drug discovery. Pharmacol Rev 66:334–395
Sukumar N, Das S (2011) Current trends in virtual high throughput screening using ligand-based and structure-based methods. Comb Chem High Throughput Screen 14:872–888
Fukunishi Y (2009) Structure-based drug screening and ligand-based drug screening with machine learning. Comb Chem High Throughput Screen 12:397–408
Maggiora G, Vogt M, Stumpfe D, Bajorath J (2014) Molecular similarity in medicinal chemistry. J Med Chem 57:3186–3204
Bender A, Mussa HY, Glen RC, Reiling S (2004) Similarity searching of chemical databases using atom environment descriptors (MOLPRINT 2D): evaluation of performance. J Chem Inf Comput Sci 44:1708–1718
Rogers D, Hahn M (2010) Extended-connectivity fingerprints. J Chem Inf Model 50:742–754
Durant JL, Leland BA, Henry DR, Nourse JG (2002) Reoptimization of MDL keys for use in drug discovery. J Chem Inf Comput Sci 42:1273–1280
Golovin A, Henrick K (2009) Chemical substructure search in SQL. J Chem Inf Model 49:22–27
Ehrlich HC, Henzler AM, Rarey M (2013) Searching for recursively defined generic chemical patterns in nonenumerated fragment spaces. J Chem Inf Model 53:1676–1688
Caporuscio F, Tafi A (2011) Pharmacophore modelling: a forty year old approach and its modern synergies. Curr Med Chem 18:2543–2553
Güner OF, Bowen JP (2014) Setting the record straight: the origin of the pharmacophore concept. J Chem Inf Model 54:1269–1283
Horvath D (2011) Pharmacophore-based virtual screening. In: Bajorath J (ed) Chemoinformatics and computational chemical biology. Humana Press, Totowa, pp 261–298. doi:10.1007/978-1-60761-839-3_11
Nicholls A, McGaughey GB, Sheridan RP, Good AC, Warren G, Mathieu M, Muchmore SW, Brown SP, Grant JA, Haigh JA, Nevins N, Jain AN, Kelley B (2010) Molecular shape and medicinal chemistry: a perspective. J Med Chem 53:3862–3886
Finn PW, Morris GM (2013) Shape-based similarity searching in chemical databases. Wiley Interdiscip Rev Comput Mol Sci 3:226–241
Hawkins PC, Skillman AG, Nicholls A (2007) Comparison of shape-matching and docking as virtual screening tools. J Med Chem 50:74–82
Armstrong MS, Morris G, Finn P, Sharma R, Moretti L, Cooper R, Richards WG (2010) ElectroShape: fast molecular similarity calculations incorporating shape, chirality and electrostatics. J Comput Aided Mol Des 24:789–801
Vainio MJ, Puranen JS, Johnson MS (2009) ShaEP: molecular overlay based on shape and electrostatic potential. J Chem Inf Model 49:492–502
Berenger F, Voet A, Lee X, Zhang K (2014) A rotation–translation invariant molecular descriptor of partial charges and its use in ligand-based virtual screening. J Cheminform 6:23
Nicholls A, Grant JA (2005) Molecular shape and electrostatics in the encoding of relevant chemical information. J Comput Aided Mol Des 19:661–686
Simonin C, Awale M, Brand M, van Deursen R, Schwartz J, Fine M, Kovacs G, Hafliger P, Gyimesi G, Sithampari A, Charles RP, Hediger MA, Reymond JL (2015) Optimization of TRPV6 calcium channel inhibitors using a 3D ligand-based virtual screening method. Angew Chem Int Ed Engl 54:14748–14752
Chen Y, Liu ZL, Fu TM, Li W, Xu XL, Sun HP (2015) Discovery of new acetylcholinesterase inhibitors with small core structures through shape-based virtual screening. Bioorg Med Chem Lett 25:3442–3446
Hevener KE, Mehboob S, Su PC, Truong K, Boci T, Deng J, Ghassemi M, Cook JL, Johnson ME (2012) Discovery of a novel and potent class of F. tularensis enoyl-reductase (FabI) inhibitors by molecular shape and electrostatic matching. J Med Chem 55:268–279
Kumar A, Ito A, Hirohama M, Yoshida M, Zhang KY (2016) Identification of new SUMO activating enzyme 1 inhibitors using virtual screening and scaffold hopping. Bioorg Med Chem Lett 26:1218–1223
Kumar A, Ito A, Takemoto M, Yoshida M, Zhang KY (2014) Identification of 1,2,5-oxadiazoles as a new class of SENP2 inhibitors using structure based virtual screening. J Chem Inf Model 54:870–880
Wiggers HJ, Rocha JR, Fernandes WB, Sesti-Costa R, Carneiro ZA, Cheleski J, da Silva AB, Juliano L, Cezari MH, Silva JS, McKerrow JH, Montanari CA (2013) Non-peptidic cruzain inhibitors with trypanocidal activity discovered by virtual screening and in vitro assay. PLoS Negl Trop Dis 7:e2370
Kaserer T, Rigo R, Schuster P, Alcaro S, Sissi C, Schuster D (2016) Optimized virtual screening workflow for the identification of novel G-Quadruplex ligands. J Chem Inf Model 56:484–500
Kumar A, Zhang KY (2016) Application of shape similarity in pose selection and virtual screening in CSARdock2014 exercise. J Chem Inf Model 56:965–973
Anighoro A, Bajorath J (2016) Three-dimensional similarity in molecular docking: prioritizing ligand poses on the basis of experimental binding modes. J Chem Inf Model 56:580–587
Kelley BP, Brown SP, Warren GL, Muchmore SW (2015) POSIT: flexible shape-guided docking for pose prediction. J Chem Inf Model 55:1771–1780
Wu G, Vieth M (2004) SDOCKER: a method utilizing existing X-ray structures to improve docking accuracy. J Med Chem 47:3142–3148
Fukunishi Y, Nakamura H (2008) Prediction of protein–ligand complex structure by docking software guided by other complex structures. J Mol Graph Model 26:1030–1033
Fukunishi Y, Nakamura H (2012) Integration of ligand-based drug screening with structure-based drug screening by combining maximum volume overlapping score with ligand docking. Pharmaceuticals (Basel) 5:1332–1345
Huang SY, Li M, Wang J, Pan Y (2016) HybridDock: a hybrid protein–ligand docking protocol integrating protein- and ligand-based approaches. J Chem Inf Model 56:1078–1087
Roy A, Srinivasan B, Skolnick J (2015) PoLi: a virtual screening pipeline based on template pocket and ligand similarity. J Chem Inf Model 55:1757–1770
Kumar A, Zhang KY (2016) A pose prediction approach based on ligand 3D shape similarity. J Comput Aided Mol Des 30:457–469
Damm-Ganamet KL, Smith RD, Dunbar JB Jr, Stuckey JA, Carlson HA (2013) CSAR benchmark exercise 2011–2012: evaluation of results from docking and relative ranking of blinded congeneric series. J Chem Inf Model 53:1853–1870
Dunbar JB Jr, Smith RD, Damm-Ganamet KL, Ahmed A, Esposito EX, Delproposto J, Chinnaswamy K, Kang YN, Kubish G, Gestwicki JE, Stuckey JA, Carlson HA (2013) CSAR data set release 2012: ligands, affinities, complexes, and docking decoys. J Chem Inf Model 53:1842–1852
Smith RD, Dunbar JB Jr, Ung PM, Esposito EX, Yang CY, Wang S, Carlson HA (2011) CSAR benchmark exercise of 2010: combined evaluation across all submitted scoring functions. J Chem Inf Model 51:2115–2131
Geballe MT, Skillman AG, Nicholls A, Guthrie JP, Taylor PJ (2010) The SAMPL2 blind prediction challenge: introduction and overview. J Comput Aided Mol Des 24:259–279
Mobley DL, Liu S, Lim NM, Wymer KL, Perryman AL, Forli S, Deng N, Su J, Branson K, Olson AJ (2014) Blind prediction of HIV integrase binding from the SAMPL4 challenge. J Comput Aided Mol Des 28:327–345
Skillman AG (2012) SAMPL3: blinded prediction of host-guest binding affinities, hydration free energies, and trypsin inhibitors. J Comput Aided Mol Des 26:473–474
Carlson HA, Smith RD, Damm-Ganamet KL, Stuckey JA, Ahmed A, Convery MA, Somers DO, Kranz M, Elkins PA, Cui G, Peishoff CE, Lambert MH, Dunbar JB Jr (2016) CSAR 2014: a benchmark exercise using unpublished data from pharma. J Chem Inf Model 56:1063–1077
Kumar A, Zhang KY (2013) Investigation on the effect of key water molecules on docking performance in CSARdock exercise. J Chem Inf Model 53:1880–1892
Kumar A, Zhang KY (2012) Computational fragment-based screening using RosettaLigand: the SAMPL3 challenge. J Comput Aided Mol Des 26:603–616
Voet AR, Kumar A, Berenger F, Zhang KY (2014) Combining in silico and in cerebro approaches for virtual screening and pose prediction in SAMPL4. J Comput Aided Mol Des 28:363–373
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28:235–242
Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242
Krissinel E, Henrick K (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60:2256–2268
Hawkins PC, Nicholls A (2012) Conformer generation with OMEGA: learning from the data set and the analysis of failures. J Chem Inf Model 52:2919–2936
Hawkins PC, Skillman AG, Warren GL, Ellingson BA, Stahl MT (2010) Conformer generation with OMEGA: algorithm and validation using high quality structures from the protein databank and Cambridge structural database. J Chem Inf Model 50:572–584
OMEGA 2.5.1.4: OpenEye Scientific Software, Santa Fe. http://www.eyesopen.com
Hawkins PCD, Skillman AG, Nicholls A (2006) Comparison of shape-matching and docking as virtual screening tools. J Med Chem 50:74–82
ROCS 3.2.0.4: OpenEye Scientific Software, Santa Fe. http://www.eyesopen.com
Rogers DJ, Tanimoto TT (1960) A computer program for classifying plants. Science 132:1115–1118
Fleishman SJ, Leaver-Fay A, Corn JE, Strauch EM, Khare SD, Koga N, Ashworth J, Murphy P, Richter F, Lemmon G, Meiler J, Baker D (2011) RosettaScripts: a scripting language interface to the Rosetta macromolecular modeling suite. PLoS One 6:e20161
Bower MJ, Cohen FE, Dunbrack RL Jr (1997) Prediction of protein side-chain rotamers from a backbone-dependent rotamer library: a new homology modeling tool. J Mol Biol 267:1268–1282
Dunbrack RL Jr, Karplus M (1993) Backbone-dependent rotamer library for proteins. Application to side-chain prediction. J Mol Biol 230:543–574
Li Z, Scheraga HA (1987) Monte Carlo-minimization approach to the multiple-minima problem in protein folding. Proc Natl Acad Sci USA 84:6611–6615
Warren GL, Andrews CW, Capelli AM, Clarke B, LaLonde J, Lambert MH, Lindvall M, Nevins N, Semus SF, Senger S, Tedesco G, Wall ID, Woolven JM, Peishoff CE, Head MS (2006) A critical assessment of docking programs and scoring functions. J Med Chem 49:5912–5931
Plewczynski D, Lazniewski M, Augustyniak R, Ginalski K (2011) Can we trust docking results? Evaluation of seven commonly used programs on PDBbind database. J Comput Chem 32:742–755
Good AC, Liu J, Hirth B, Asmussen G, Xiang Y, Biemann HP, Bishop KA, Fremgen T, Fitzgerald M, Gladysheva T, Jain A, Jancsics K, Metz M, Papoulis A, Skerlj R, Stepp JD, Wei RR (2012) Implications of promiscuous Pim-1 kinase fragment inhibitor hydrophobic interactions for fragment-based drug design. J Med Chem 55:2641–2648
Acknowledgments
We acknowledge the Hokusai Great Wave supercomputer at RIKEN for the supercomputing resources used in this study. We acknowledge RIKEN Pioneering Project in Dynamic Structural Biology for funding. We thank members of our lab for help and discussions.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kumar, A., Zhang, K.Y.J. Prospective evaluation of shape similarity based pose prediction method in D3R Grand Challenge 2015. J Comput Aided Mol Des 30, 685–693 (2016). https://doi.org/10.1007/s10822-016-9931-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-016-9931-2