Abstract
Purpose
To perform a preliminary exploration of a new embryo rank in clinical practice by combining the embryo chromosome copy number and mitochondrial copy number analysis of DNA extracted from embryo culture medium and blastocoel fluid.
Method
Eighty-three ICSI embryos from day 2 or day 3 were cultured to day 5 or day 6. Thirty-two blastocysts of 3 cc or above were obtained. Culture medium and blastocoel fluid were collected at 24 h before blastocyst formation. The genomic DNA and mitochondrial DNA (mtDNA) from the culture medium combined with blastocoel fluid and the whole blastocyst were amplified and sequenced by MALBAC-NGS. We compared the chromosomal information generated by the new protocol from the culture medium and the information employed by the whole embryo method. A multivariable linear regression was performed to study the impact of the blastocyst morphological score, chromosomal abnormality, embryo mtDNA copy number, and female age on the culture medium mtDNA copy number.
Results
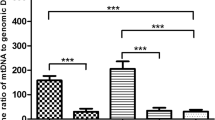
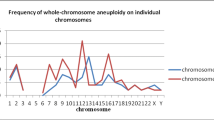
(1) The DNA from 31 blastocysts was successfully amplified, and the successful amplification rate was 96.9% (31/32). The success rate of the amplification of genomic DNA extracted from the culture medium was 87.5% (28/32). (2) There were 18 blastocysts in which the less invasive method and the whole embryo method revealed the same results. The consistency rate was 66.7% (18/27). (3) The culture medium mitochondrial DNA copy number (MCN) had a significantly positive correlation with the blastocyst mitochondrial DNA copy number (P = 0.001), female age (P = 0.012), and blastocyst score (P = 0.014), but there was no obvious correlation with blastocyst chromosome (P = 0.138).
Conclusions
The preliminary exploration result of the less invasive approach for having an embryo rank was not satisfying, which still awaits further long-term evaluation.




Similar content being viewed by others
References
Kushnir VA, Darmon SK, Albertini DF, et al. Effectiveness of in vitro fertilization with preimplantation genetic screening: a reanalysis of United States assisted reproductive technology data 2011-2012 [J]. Fertil Steril. 2016;106(1):75–9.
Schoolcraft WB, Treff NR, Stevens JM, et al. Live birth outcome with trophectoderm biopsy, blastocyst vitrification, and single-nucleotide polymorphism microarray-based comprehensive chromosome screening in infertile patients [J]. Fertil Steril. 2011;96(3):638–40.
Scottrt JR, Upham KM, Forman EJ, et al. Blastocyst biopsy with comprehensive chromosome screening and fresh embryo transfer significantly increases in vitro fertilization implantation and delivery rates: a randomized controlled trial [J]. Fertil Steril. 2013;100(3):697–703.
Tobler KJ, Brezina PR, Benner AT, du L, Xu X, Kearns WG. Two different microarray technologies for preimplantation genetic diagnosis and screening, due to reciprocal translocation imbalances, demonstrate equivalent euploidy and clinical pregnancy rates [J]. J Assist Reprod Genet. 2014;31(7):843–50.
Caglar GS, Asimakopoulos B, Nikolettos N, et al. Preimplantation genetic diagnosis for aneuploidy screening in repeated implantation failure[J]. Reprod BioMed Online. 2005;10(3):381–8.
Reisesilva AR, Bruno C, Fleurot R, et al. Alteration of DNA demethylation dynamics by in vitro culture conditions in rabbit pre-implantation embryos [J]. Epigenetics. 2012;7(5):440–6.
Frenandez-Gonzalez R, Ramirez MA, Pericuesta E, et al. Histone modifications at the blastocyst Axin1(Fu) locus mark the heritability of in vitro culture-induced epigenetic alterations in mice [J]. Biol Reprod. 2010;83(5):720–7.
Market-Velker BA, Fernandes AD, Mann MR. Side-by-side comparison of five commercial media systems in a mouse model: suboptimal in vitro culture interferes with imprint maintenance [J]. Biol Reprod. 2010;83(6):938–50.
Yu Y, Zhao Y, Li R, Li L, Zhao H, Li M, et al. Assessment of the risk of blastomere biopsy during preimplantation genetic diagnosis in a mouse model: reducing female ovary function with an increase in age by proteomics method [J]. J Proteome Res. 2013;12(12):5475–86.
Zhao HC, Zhao Y, Li M, et al. Aberrant epigenetic modification in murine brain tissues of offspring from preimplantation genetic diagnosis blastomere biopsies [J]. Biol Reprod. 2013;89(5):117.
Dawson A, Griesinger G, Diedrich K. Screening oocytes by polar body biopsy[J]. Reprod BioMed Online. 2006;13:104–9.
Palini S, Galluzzi L, de Stefani S, Bianchi M, Wells D, Magnani M, et al. Genomic DNA in human blastocoele fluid - Reproductive BioMedicine Online. Reprod BioMed Online. 2013;26:603–10.
Hammond ER, McGillivray BC, Wicker SM, et al. Characterizing nuclear and mitochondrial DNA in spent embryo culture media: genetic contamination identified [J]. Fertil Steril. 2017;107(1):220–228.e5.
Stigliani S, Anserini P, Venturini PL, Scaruffi P. Mitochondrial DNA content in embryo culture medium is significantly associated with human embryo fragmentation [J]. Hum Reprod. 2013;28(10):2652–60.
Gianaroli L, et al. Blastocentesis: a source of DNA for preimplantation genetic testing. Results from a pilot study. Fertil Steril. 2014;102:1692–9.
Gardner DK, Schoolcraft WB. Culture and transfer of human blastocysts. Curr Opin Obstet Gynecol. 1999;11:307.
Gardner DK, Lane M, Stevens J, Schoolcraft WB. Changing the start temperature and cooling rate in a slow-freezing protocol increases human blastocyst viability. Fertil Steril. 2003;79:407–10.
Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–6.
Belandres D, Shamonki M, Arrach N. Current status of spent embryo media research for preimplantation genetic testing. J Assist Reprod Genet. 2019;36:819–26. https://doi.org/10.1007/s10815-019-01437-6.
Wu H, Ding C, Shen X, Wang J, Li R, Cai B, et al. Medium-based noninvasive preimplantation genetic diagnosis for human alpha-thalassemias-SEA [J]. Medicine (Baltimore). 2015;94(12):e669.
Gianaroli L, Magli MC, Pomante A, Crivello AM, Cafueri G, Valerio M, et al. Blastocentesis: a source of DNA for preimplantation genetic testing. Results from a pilot study [J]. Fertil Steril. 2014;102(6):1692–9 e1696.
Ho JR, Arrach N, Rhodes-Long K, Ahmady A, Ingles S, Chung K, et al. Pushing the limits of detection: investigation of cell-free DNA for aneuploidy screening in embryos. Fertil Steril. 2018;110(3):467–75.
Xu J, Fang R, Chen L, et al. Noninvasive chromosome screening of human embryos by genome sequencing of embryo culture medium for in vitro fertilization [J]. Proc Natl Acad Sci U S A. 2016;113(42):11907–12.
Feichtinger M, Vaccari E, Carli L, Wallner E, Mädel U, Figl K, et al. Non-invasive preimplantation genetic screening using array comparative genomic hybridization on spent culture media: a proof-of-concept pilot study. Reprod BioMed Online. 2017;34(6):583–9.
Vera-Rodriguez M, Diez-Juan A, Jimenez-Almazan J, Martinez S, Navarro R, Peinado V, et al. Origin and composition of cell-free DNA in spent medium from human embryo culture during preimplantation development. Hum Reprod. 2018;33(4):745–56.
Chan KC, Jiang P, Sun K, Cheng YK, Tong YK, Cheng SH, et al. Second generation noninvasive fetal genome analysis reveals de novo mutations, single-base parental inheritance, and preferred DNA ends. Proc Natl Acad Sci U S A. 2016;50:E8159–68.
Munné S, et al. Detailed investigation into the cytogenetic constitution and pregnancy outcome of replacing mosaic blastocysts detected with the use of high-resolution next-generation sequencing. Fertil Steril. 2017;108:62.
Fragouli E, et al. Cytogenetic analysis of human blastocysts with the use of FISH, CGH and aCGH: Scientific data and technical evaluation. Hum Reprod. 2011;26:480.
Ruttanajit T, et al. Detection and quantitation of chromosomal mosaicism in human blastocysts using copy number variation sequencing. Prenat Diagn. 2015;36:154.
Van EJ, et al. Chromosomal mosaicism in human preimplantation embryos: a systematic review. Hum Reprod Update. 2011;17:620–7.
Vera-Rodriguez M, Rubio C. Assessing the true incidence of mosaicism in preimplantation embryos. Fertil Steril. 2017;107:1107.
Liu J, et al. DNA microarray reveals that high proportions of human blastocysts from women of advanced maternal age are aneuploid and mosaic. Biol Reprod. 2012;87:148.
Fragouli E, Alfarawati S, Spath K, Babariya D, Tarozzi N, Borini A, et al. Analysis of implantation and ongoing pregnancy rates following the transfer of mosaic diploid-aneuploid blastocysts [J]. Hum Genet. 2017;136(7):805–19.
Greco E, Minasi MG, Fiorentino F. Healthy Babies after Intrauterine Transfer of Mosaic Aneuploid Blastocysts [J]. N Engl J Med. 2015;373(21):2089–90.
Taylor TH, GitlinS A, Patrick JL, et al. The origin, mechanisms, incidence and clinical consequences of chromosomal mosaicism in humans [J]. Hum Reprod Update. 2014;20(4):571–81.
Werner MD, Hong KH, Franasiak JM, Forman EJ, Reda CV, Molinaro TA, et al. Sequential versus Monophasic Media Impact Trial (SuMMIT): a paired randomized controlled trial comparing a sequential media system to a monophasic medium. Fertil Steril. 2016;105:1215–21. https://doi.org/10.1016/j.fertnstert.2016.01.005.
Kuznyetsov V, Madjunkova S, Antes R, Abramov R, Motamedi G, Ibarrientos Z, et al. Evaluation of a novel non-invasive preimplantation genetic screening approach. PLoS One. 2018;13(5):e0197262.
Mukaida T, Oka C, Goto T, Takahashi K. Artificial shrinkage of blastocoeles using either a micro-needle or a laser pulse prior to the cooling steps of vitrification improves survival rate and pregnancy outcome of vitrified human blastocysts. Hum Reprod. 2006;21(12):3246–52.
Chan C, Liu V, Lau E, Yeung WS, Ng EH, Ho PC. Mitochondrial DNA content and 4977 bp deletion in unfertilized oocytes. Mol Hum Reprod. 2005;11:843–6.
Van Blerkom J. Mitochondrial function in the human oocyte and embryo and their role in developmental competence. Mitochondrion. 2011;11:797–813.
Shang W, et al. Comprehensive chromosomal and mitochondrial copy number profiling in human IVF embryos. Reprod BioMed Online. 2017. https://doi.org/10.1016/j.rbmo.2017.10.110.
Fragouli E, Wells D. Mitochondrial DNA assessment to determine oocyte and embryo viability. Semin Reprod Med. 2015;33(6):401–9.
Acknowledgments
We are grateful to our patients for their sincere participation in this study and thank each author for their hard work.
Authors’ roles
X.B. designed and coordinated the study. H.X., C.Y., L.F., and X.S. collected the clinical cases. J.Z. and H.X. performed and analyzed the data. J.Z. wrote the draft of the manuscript. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval was obtained from the Scientific and Ethical Committee of Tianjin Medical University for the experimental protocol (IRB2017-155-01). A detailed patient consent form was signed for each embryo sample used in this study.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, J., Xia, H., Chen, H. et al. Less-invasive chromosome screening of embryos and embryo assessment by genetic studies of DNA in embryo culture medium. J Assist Reprod Genet 36, 2505–2513 (2019). https://doi.org/10.1007/s10815-019-01603-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-019-01603-w