Abstract
The successful development of microalgae-based biofuel production will rely on improvements in the amount and rate of fuel molecule precursor accumulation. Mutagenesis and selection is an attractive approach to improve fuel molecule productivity. Previous screening methods have been laborious, the numbers of mutants isolated have been small, and overall performance of mutants may have been compromised by the presence of deleterious secondary mutations generated by random mutagenesis that affect other cellular processes and growth. We report an improved method of isolating high triacylglycerol (TAG) accumulating mutants of a Chlorella sp., KAS603, using flow cytometric-based selection. In addition to selection for high TAG accumulating strains, the method requires that growth of mutants be competitive with other cells in the population. Not only is growth competitive, but there is improved performance in TAG accumulation with repeated selection, suggesting purification from deleterious secondary mutations. The procedure resulted in the isolation of mutants with far higher efficiency (thousands of fold) that outperformed wild type substantially better (1.8–2.5-fold) than with previous methods. This opens the door to new approaches to the characterization of genes involved in TAG accumulation and other cellular processes.
Similar content being viewed by others
Introduction
As research and technology for microalgae-based biofuels develops, it is clear that lowering the cost of production is the single most important factor for commercial viability. There are many factors associated with large-scale production, ranging from the biology to engineering of culture systems, nutrient regime in the growth medium, harvesting and extraction processes, etc., and different aspects contribute differently to the overall cost. Analyses indicate that the two biggest components towards reducing cost would be improved lipid content per cell (for lipid-based fuel precursor molecules) and improved algae growth and biomass accumulation (Pienkos et al. 2011). Although many native algae species have attractive growth and lipid accumulation characteristics, their performance under outdoor cultivation conditions can be less relative to laboratory conditions. In addition, even though native species may accumulate adequate amounts of lipid or triacylglycerol (TAG), the rate of accumulation may hinder economical production. Microalgae species that accumulate fuel precursors in abundance, at a rapid rate, will be most desirable for production systems.
There are two general approaches to improve lipid accumulation ability: selection of better performing strains and directed genetic manipulation. The latter is the “rational” approach, in which specific target genes suspected to be important are manipulated to improve fuel precursor production characteristics. Its major advantage is that the gene to be manipulated is known. Its disadvantages are that (1) it may not be clear until the manipulation was complete whether the gene was a good target, (2) the approach may not encompass genes that have an unanticipated beneficial effect (e.g., transcription factors that regulate pathways may be difficult to identify), and (3) because the manipulation leads to a genetically modified organism, regulatory issues come into play for production. Selection approaches have proven valuable in isolating environmental strains with improved production characteristics (Aquatic Species Program Closeout Report 1998; Mutanda et al. 2011). Selection approaches can be applied to wild-type strains (Montero et al. 2011) or be combined with mutagenesis as a means of genetic improvement with the advantage that no foreknowledge of the genes involved is needed. This enables identification of both anticipated and unanticipated genes responsible for improved phenotypes, and mutagenized and/or selected organisms are not classified as genetically modified. The drawbacks of mutagenesis and selection approaches are that (1) a specific selection method must be available for the desired phenotype; (2) in the case of random mutagenesis, multiple secondary mutations are generated in the genome, some of which could be detrimental to growth or productivity; and (3) identification of the locus of the mutation responsible for the desired phenotype can be challenging given the fact that it may only result from a point mutation in a background of numerous point mutations in other genes. However, with modern techniques of inexpensive high-throughput sequencing available, reasonable strategies can be devised to identify the responsible mutation.
Previous work has demonstrated the utility of selecting algal mutants for improved productivity. In chlorophyte algae, a phenotypic screen for inhibited starch accumulation has resulted in isolation of “starchless” mutants (Ball et al. 1991; Delrue et al. 1992; Plumed et al. 1996; Zabawinski et al. 2001). Further characterization of such a mutant in Chlamydomonas reinhardtii demonstrated substantially improved TAG accumulation ability under nitrogen limitation, with a 2-fold faster rate and 2-fold higher accumulation than wild type (Wang et al. 2009). These approaches have relied on direct plating of mutagenized cells followed by screening of large numbers (tens of thousands) of mutants to identify the desired phenotype, which although effective, is laborious. Another phenotype isolated by plate screening was the small chlorophyll antenna size mutants of the diatom Cyclotella cryptica (Huesemann et al. 2009). These mutants performed well in the lab, but not so in outdoor conditions (Huesemann et al. 2009). The reason for poor performance was not elucidated, but one possibility could have been the presence of detrimental secondary mutations. More recently, procedures were developed for Nannochloropsis sp. to select for higher lipid accumulating mutants using fluorescence-based staining of lipid droplets coupled with flow cytometric selection of the highest percentage of lipid containing cells (Doan and Obbard 2011, 2012). Substantial improvements in lipid content were obtained, especially after repeated rounds of selection (Doan and Obbard 2012). The process was more efficient and less labor intensive than plate screening and resulted in isolation of 13 mutants out of thousands screened. Two mutants demonstrated a 1.5-fold improvement in fatty acid methyl ester (FAME) content relative to wild type (Doan and Obbard 2012). Nannochloropsis is a member of the stramenopile algae and is a promising production strain (Hu et al. 2008), so the strains that were most improved may have a practical application.
The chlorophytes are an evolutionarily distinct class of microalgae with promising production characteristics (Hu et al. 2008). We have worked with Chlorella sp. KAS603, a proprietary species isolated by Kuehnle AgroSystems (Honolulu, Hawaii, USA) with attractive growth, environmental flexibility, and TAG accumulation characteristics. This species accumulates TAG rapidly in response to combined nitrogen and phosphorous depletion. In this report, we systematically develop a mutagenesis and selection scheme using flow cytometric sorting to isolate mutants with improved TAG accumulation characteristics. The data suggest that although epigenetic variation occurs, stable phenotypes are consistent with genetic alterations. We demonstrate a much more efficient recovery of high lipid accumulation mutants with higher TAG levels than with previously reported approaches. These results also open the door towards new approaches to characterize genes involved in TAG accumulation or other cellular processes.
Materials and methods
Algal strains and culture conditions
The green alga Chlorella sp. KAS603 (a proprietary strain from Kuehnle AgroSystems) was grown in 125-mL stoppered Erlenmeyer flasks containing 50 mL 14G growth medium (General Atomics 2012) at 25 °C under 500 μmol photons m−2 s−1 continuous light. Cells were inoculated into 14G growth medium starting with 2 × 106 cells mL−1. Individual sorted cells were grown in 24-well plates before further cultivation in 125 mL Erlenmeyer flasks. To induce TAG accumulation, cells were placed in induction medium (14 L) lacking nitrate and phosphate (General Atomics 2012). Cell growth was measured by counting cell number using a hemocytometer. This species is robust and massively scalable and has been used to produce over one million liters of culture in open ponds for biofuel purposes (A. Kuehnle, personal communication).
Mutagenesis methods
UV mutagenesis was performed according to Ball et al. (1991). Briefly, exponentially grown cells were harvested, washed, and suspended in 14G medium at 5 × 106 cells mL−1. Cells were placed in a 60-mm petri dish covered with two layers of a Kimwipe and irradiated using a UV cross linker (UVP CX-2000) at the maximum intensity (1 joule cm−2) at a distance of 7 cm for 30 sec. Irradiated cells were allowed to recover in the dark for 24 h. Cells were then either inoculated into liquid, or plated on agar plates, and placed at 25 °C under 500 μmol photons m−2 s−1 continuous light for 5–7 days or until colonies were visible. Fifty percent survival was considered as optimal irradiance for mutant generation. For double mutagenesis, two singly mutagenized cell lines were subjected to the same treatment.
Chemically mutagenized cells were treated with ethyl methansulfonate (EMS, Sigma); 5 × 108 cells mL−1 were incubated in 0.28 M EMS in phosphate buffer, pH 7 in the dark at 25 °C for various time intervals with constant shaking. The reaction was stopped by adding 5 % sodium thiosulfate at 15, 30, 45, 60, 75, and 90 min. Samples were washed twice with phosphate buffer and resuspended into the medium. Mutagenized cells were plated onto agar plates to determine viability and also grown in liquid medium for sorting by flow cytometry. Untreated cells were used as a control. We selected 45 min of mutagenesis, which resulted in 50 % survival.
Characterization of triacylglycerol content by BODIPY fluorescence using imaging flow cytometry
TAG accumulation was monitored using BODIPY fluorescence as proxy using an Amnis ImageStream imaging flow cytometer. At different times during the course of an experiment, 1 × 107 cells were removed from a culture and pelleted and stored at −20 °C. For analysis, pellets were thawed, resuspended in potassium phosphate buffer (0.1 M, pH 7), and stained with 2.6 μg mL−1 BODIPY (4,4-difluoro-3a,4a-diaza-s-indacene) (Invitrogen, USA) by incubating for 20 min at room temperature. After 20 min, samples were run through the ImageStream. Typically, data were collected on 10,000–20,000 cells. Data were analyzed using IDEAS™ software for the ImageStream and plotted as chlorophyll vs. BODIPY fluorescence.
Starch screening
Six to ten million (6–10 × 106) cells of a particular mutant were harvested, resuspended in a microcentrifuge tube containing 1.2 mL methanol, vortexed vigorously for 1 min, and then centrifuged. If pellets were still green, 1.2 mL methanol was added and heated at 80 °C for a few hours until the green color disappeared (alternatively, pellets were extracted with a higher volume of methanol). Pellets were dried and iodine/KI (5 mM iodine/10 mM KI) solution was added. After 2 min of incubation, cells containing starch changed to a dark-brown or black color while cells containing less starch changed to a pale yellow color.
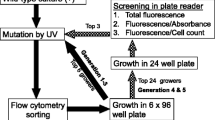
Mutant screening by flow cytometer sorting
Cells (2–3 × 107 cells mL−1) were harvested and stained with BODIPY as described above. TAG level was measured using BODIPY fluorescence as a proxy (Cooper et al. 2010). Cells were excited with a 488-nm laser, and emission was evaluated at 530/40 nm on a Becton Dickinson Influx sorting flow cytometer. Different isolation parameters were tested (see “Results”), but the only top few percent of cells in terms of BODIPY fluorescence were isolated. We routinely sorted 10,000 cells to be used for plating and 100,000–200,000 cells for growth in the liquid medium. Cells were plated immediately after sorting and cells grown in liquid culture were used for resorting.
Results
Imaging flow cytometry analysis of TAG accumulation
We examined the TAG accumulation process resulting from N and P limitation on wild-type cells using imaging flow cytometry as shown in Fig. 1. Imaging flow cytometry using the Amnis ImageStream X enables not only scatter plot analyses typical of a sorting flow cytometer, but high-resolution microscopic imaging of all cells in the population. Based on the microscopic appearance of cells (Fig. 1b), we assigned them to four different subpopulations: (1) low lipid, in which very little BODIPY fluorescence was visible; (2) intermediate lipid, with intermediate amounts of BODIPY fluorescence; (3) high lipid with chlorophyll (cells containing a maximum amount of BODIPY fluorescence and still containing chlorophyll); and (4) high lipid with no chlorophyll (cells containing a maximum amount of BODIPY fluorescence that lacked chlorophyll). Repeated examination from several experiments revealed a general robustness to the classification scheme, although cells near borders between categories can exhibit intermediate appearances. The classification scheme allows a consistent evaluation to be made from experiment to experiment. These data contain additional information than a bulk measurement because they enable visualization of the spread in the data at individual time points and subpopulations that differ in one parameter but not in another (e.g., high lipid but with or without chlorophyll). Even though the spread in the population can be substantial, because large numbers of cells are being imaged (10–20,000), the standard error is small, and the means are realistic assessments. These concepts are demonstrated in panels c and d of Fig. 1, which show the means plotted with the standard deviation (Fig. 1c) or the standard error (Fig. 1d). The time course shows an increasing accumulation of lipid over time, with a loss of chlorophyll at the latest time (Fig. 1). This pattern has been repeatedly observed in this species and in other classes of algae (Traller and Hildebrand, unpublished data) and may be a general phenomenon.
Imaging flow cytometry analysis of TAG accumulation in wild-type cells resulting from N and P limitation. a Scatter plots of chlorophyll and BODIPY fluorescence on different days after placing cells in induction medium. Five distinct subpopulations of cells were identified based on their microscopic appearance, which are delineated by different colored gates. b Representative images from four of the five subpopulations (the fifth was dead cells or debris). Subpopulations are 1 low lipid, 2 intermediate lipid, 3 high lipid with chlorophyll, and 4 high lipid with no chlorophyll. c, d Plot of means of BODIPY fluorescence. Error bars represent standard deviation (c) or standard error (d)
TAG accumulation ability is related to the stage of culture growth
To screen possible mutants for improved TAG accumulation, consistency in the response is required, so we investigated the relationship between culture growth status and TAG accumulation ability upon subsequent N and P limitation. During a growth experiment with wild-type cells, on five successive days spanning exponential growth into stationary phase, aliquots of cells were removed from the culture and placed in induction medium, following which TAG level was monitored by BODIPY fluorescence over 3 days (Fig. 2). Growth rate showed no significant differences between duplicate wild-type cultures (Fig. 2a). There was a consistent decrease in subsequent TAG accumulation with successive days of culturing, with the exception of day 5 when there was a substantial increase in TAG between days 2 and 3 of induction (Fig. 2b). By averaging the mean BODIPY fluorescence for both cultures over all three induction days, we monitored a general trend of reduction of TAG accumulation ability as the cells entered the stationary phase, but with prolonged stationary conditions, the delayed response with a high level of TAG accumulation on the last day of monitoring increased the average level on day 5 (Fig. 2c). These data suggest longer term effects of culture history over the subsequent ability to accumulate TAG. The reasons for this are unclear and beyond the scope of the present study; however, given that these were clonal populations of cells, one possibility is that epigenetic processes are involved. Based on these data, we chose day 2 after inoculum to harvest cells for TAG accumulation measurements for all subsequent experiments.
TAG accumulation of wild-type cells at different culture stages. a Growth curve of two wild-type cultures (WT3 and WT6) grown through exponential into stationary phase and aliquots isolated on days 1–5 of the culture. b Plots of relative changes in TAG levels from aliquots that were subjected to TAG induction under N and P limitation, monitored by BODIPY fluorescence over 3 days. c Plot of the relative change in the means of TAG accumulation over the 3-day period for WT3 and WT6 on each day isolated from the growth experiment. d Mean and average standard deviation for each time point for two wild-type replicates in eight separate TAG induction experiments
Since exact control over culture status is not possible (the condition of cells at “day x” for any given culture is not exactly the same for another), even a few hours variation at a sensitive time (e.g., between day 2 and day 3) could result in a dramatic change in TAG accumulation ability. For this reason, we used two wild-type cultures in all subsequent mutant screening experiments in an attempt to account for variation. Figure 2d shows the mean and average standard deviation for each time point for two wild-type replicates in eight separate TAG induction experiments. The average percent standard deviation of the mean was 21 % across all time points. These data indicate that although variation occurred, induction levels were consistent enough to make evaluations.
Sorting different subpopulations of cells containing different TAG content
Previous work has shown that selection of wild-type populations with higher TAG content of Tetraselmis suecica resulted in stable subpopulations with higher TAG content over many days of growth (Montero et al. 2011). To evaluate whether stable populations of KAS603 cells with different inherent ability to accumulate lipid could be selected and propagated, we subjected wild-type KAS603 to combined N and P limitation for 2 days and analyzed TAG content by BODIPY fluorescence using sorting flow cytometry (Fig. 3a). A range of different TAG levels in the BODIPY-stained population were observed, with variation on the order of 100-fold (Fig. 3a). We then sorted 10 % of the population showing high (HF) and low fluorescence (LF). Cells were subsequently analyzed for growth rate and TAG level. Both isolated populations showed similar growth rate and maximum culture density (Fig. 3b); however, HF isolates showed 4-fold higher TAG accumulation than LF under N and P limitation in induction medium (Fig. 3c). These data demonstrate that subpopulations of cells with intrinsically greater or lesser ability to accumulate TAG can be isolated. Because the starting population was genetically homogeneous, the data are consistent with epigenetic variation contributing to the differences between high and low populations.
Results of sorting different subpopulations of cells containing different lipid content. a Scatter plot of forward scatter (FSC) vs. BODIPY fluorescence from the sorting flow cytometer. Relative fluorescence units are displayed. Boxes denote gates from which high and low fluorescence subpopulations of cells were isolated by sorting. b Comparison of growth rate between the high and low populations. c TAG accumulation determined by BODIPY fluorescence in the high and low subpopulations
Cell mutagenesis and screening mutants using fluorescence-activated cell sorting
The data in Fig. 3 indicate that the phenotype of high or low lipid accumulation persisted over several generations, and data in Figs. 2 and 3 suggest that epigenetic effects could be involved. Since epigenetic effects are not expected to be stable over the long term, we attempted to make genetic changes to increase phenotypic stability and to potentially further improve lipid accumulation ability. We UV-mutagenized HF isolates and, after 2 days of recovery in liquid medium, subjected cells to TAG induction and sorted cells on the basis of high BODIPY fluorescence. Two approaches were tested: sorting of a subpopulation containing 10,000 cells with the highest 1–3 % fluorescence which were plated immediately after sorting, and sorting of a subpopulation containing 100,000–200,000 cells with the highest 10 % fluorescence which were then grown in liquid culture. Plating efficiency of immediately-sorted cells was extremely low; in a typical experiment, three to five colonies out of 10,000 would be isolated if the top 1 % fluorescent cells were sorted (survival was better if the top 3 % were isolated). This suggests that the highest lipid accumulating cells were marginally viable.
Based on literature showing that deficiencies in starch accumulation in chlorophytes can result in increased TAG accumulation (Wang et al. 2009), we examined several putative mutants for the presence of starch after growth in liquid medium. Figure 4 is a comparison of wild type and several putative mutants, showing differences in the extent of iodine staining, which is indicative of the presence (dark color) or absence (light color) of starch. Several mutants exhibited a low- or no-starch phenotype; however, we cannot conclude that these actually lack the ability to accumulate starch, but only that they did not accumulate starch under the conditions tested. Two of the apparent starchless mutants were subsequently tested for TAG accumulation and showed improved ability relative to wild type (data not shown).
Because of the low recoveries of cells plated directly after sorting, and also because that approach lacked a requirement for competitive growth, we subsequently focused on characterization of sorted cells that were allowed to recover and grow in liquid medium. These were plated 2–3 days after sorting, with good plating efficiency. Random colonies were selected from these plates to isolate clonal populations that were inoculated into liquid medium. In an effort to not limit the selected phenotype, we did not include starch screening with this approach. After culture amplification, experiments were set up to compare putative mutants relative to two wild-type cultures. Examples of two comparisons are shown in Fig. 5. In comparison A (Fig. 5, left), three of the six putative mutants showed substantially better (2-fold) TAG accumulation ability than the wild-type cultures. In comparison B (Fig. 5, right), four of the ten displayed substantial improvement over wild type, and others showed slight improvement. It is important to note that not only are higher levels of TAG accumulated in the best mutants, but the rate of TAG accumulation is faster. Comparison of fluorescence levels for wild type on day 3 shows that similar levels are achieved between 1 and 1.5 days faster in the mutants (Fig. 5).
TAG accumulation in doubly mutagenized cells
To determine whether further improvement could be made to TAG accumulation, we selected two high TAG containing mutants and subjected them to an additional round of UV mutagenesis, selection, and plating. The doubly mutagenized (Mut II) progeny were then analyzed for TAG accumulation resulting from N and P limitation using imaging cytometry. We observed that double mutagenesis did not improve TAG accumulation and actually resulted in lower TAG content than wild-type cells in most cases (Fig. 6).
Changes in TAG accumulation after sequential sorting
To test whether multiple rounds sorting of high TAG accumulating strains would result in improved TAG accumulation in subsequent generations, we repeatedly sorted wild type and singly and doubly mutagenized isolates. Three rounds of sorting were performed; after each round of sorting, cells were grown and reselected for high TAG content on a weekly basis. Sorted cells were grown in growth medium for 2 days and transferred to induction medium for 3 days, and TAG level was analyzed by imaging cytometry. Figure 7 showed that after the first, second, and third round of sorting, singly mutagenized cells improved their TAG accumulation ability by 1.2-, 1.8-, and 2.2-fold higher than wild type, respectively. For the doubly mutagenized cells, a similar trend was seen, with better TAG accumulation ability occurring with additional selections. In contrast, wild type displayed no consistent trend with repeated selection (Fig. 7).
Effect of multiple sorting on TAG accumulation of singly and doubly mutagenized cells and nonmutagenized wild-type cells. Three clonal populations of cells for each treatment were sequentially selected three times using the sorting cytometer. Plots show TAG accumulation monitored by BODIPY fluorescence in induction medium. Wild-type controls are denoted with black lines, cells selected once are denoted by open symbols, cells selected twice are denoted by crosses, and cells selected three times are denoted by closed symbols
Summary of screening results
Table 1 tabulates the results of the various screens performed in this study. For UV-irradiated single mutagenesis, we analyzed a total of 87 mutants and found that 32 accumulated 1.8–3-fold higher TAG than wild type (Table 1). Fifty-five mutants showed 1–1.5-fold higher TAG level than wild type. For doubly mutagenized cells, the percent with high TAG accumulation was substantially less than singly mutagenized cells. We also attempted chemical mutagenesis, which gave similarly poor results (Table 1). Finally, no substantially improved isolates were obtained with nonmutagenized wild-type cells, indicating that the improvements had a genetic basis and that the phenotypic changes were stable under selection.
Discussion
The cell sorting approach we applied proved to be a highly efficient method to isolate mutants with better TAG accumulation characteristics. Traditional methods have relied on plating of cells after mutagenesis, followed by screening of tens of thousands of colonies for a particular phenotype. For example, starchless mutants have been isolated at an efficiency of approximately 1:10,000 (Ball et al. 1991; Delrue et al. 1992; Plumed et al. 1996; Zabawinski et al. 2001). More recently, the efficacy of continuous selection in liquid has been demonstrated in Nannochloropsis sp. which resulted in 13 mutants with increased FAME content obtained out of thousands screened (Doan and Obbard 2012). Our results on the singly mutagenized cells indicate an even higher recovery of mutants with improved TAG accumulation ability. High TAG accumulating mutants were isolated in our approach at a frequency of 1:2.7, which is 3,700 times better than typical plate screening with far less effort. Improved recovery over that in the Nannochloropsis sp. study (Doan and Obbard 2012), which used a conceptually similar sorting approach, could be due to the mutagenesis method. We show that the recovery of chemically induced mutants in much poorer than UV-induced mutants (Table 1). In addition to improved efficiency of mutant isolation, the data suggest the possibility of differences in the extent of improvement of lipid content between the two methods. One needs to consider that different organisms are being compared as well as TAG vs. FAME accumulation; however, in comparison to wild type, the chemical mutagenesis-based approach used on Nannochloropsis sp. resulted in 11 mutants with higher FAME content than wild type, but only two with greater than 1.5-fold improvement (Doan and Obbard 2012), compared with 32 mutants obtained by UV mutagenesis in this study with 1.8–2.5-fold higher TAG accumulation than wild type, and a large number of others with lesser improvement (Table 1).
In addition to improved isolation efficiency, another consideration with this approach compared with plating approaches is the effect of mutagenesis on growth rate. With plating, there is not a strict requirement for mutagenized cells to grow competitively with wild type. Colonies may grow at different rates on a plate, but they will still be present. Mutagenized cells in our study were subjected to continuous growth in competition with other cells and were only plated at the last step after selection to isolate clones. Figure 8 shows a comparison of growth rate of two clones each of singly and doubly mutagenized cells in comparison with wild type. Growth and maximum cell density is comparable in all cases.
Although intracellular TAG content has been analyzed previously using fluorescent neutral lipid stains coupled with flow cytometry (Doan and Obbard 2012), the application of imaging flow cytometry provides additional benefits. Primarily, distinct stages in the TAG accumulation process could be identified and correlated directly with the appearance of the cells (Fig. 1). The ability to image tens of thousands of cells in a short time provides statistical robustness to the analyses. This is especially important given the spread in TAG content comparing cells in the population (Fig. 1). Such a spread, on the order of up to a hundred fold, has been seen in other microalgae (Doan and Obbard 2012 and unpublished observations). Because these are clonal populations of cells, a probable explanation is that there is epigenetic variation. The results of sorting nonmutagenized cells in Fig. 3 are consistent with this.
The relation between culture stage and TAG accumulation ability, which could also relate to epigenetic variation, is shown in Fig. 2. Previous work on Nannochloropsis sp. showed a similar relationship, but those experiments monitored differences between exponential, mid-stationary, and late stationary phases, with 8- and 12-day gaps between exponential and mid-stationary and mid-stationary and late stationary phases, respectively (Doan and Obbard 2012). The results in Fig. 2 demonstrate substantial differences on the order of individual days for KAS603. In addition, as the cells enter stationary phase, there was an increase in FAME with Nannochloropsis sp. (Doan and Obbard 2012), but a sequential decrease in KAS603 with the exception of day 5 (Fig. 2). The data for day 5 suggest that the response can be complex and that there are lingering effects related to culture stage. Elucidating the mechanisms involved would be beneficial, because the rate of TAG accumulation between days 2 and 3 in this condition is the single highest rate comparing all conditions.
The most efficient approach for selecting mutants resulted from a single round of mutagenesis (Table 1). The lack of improvement in doubly mutagenized cells may likely be a result of accumulation of deleterious secondary mutations; in this case, selection was unable to rescue the high TAG accumulation phenotype (Fig. 6).
Previous work showed that successive rounds of selection of a bulk population of cells improved the overall TAG accumulation response of the bulk population of cells (Doan and Obbard 2012). In our study, we performed successive rounds of selection on individual clones of mutants (Fig. 7). For singly and doubly mutagenized cells, there was a trend of improvement of TAG accumulation ability with successive sorting (Fig. 7). Coupled with the lack of this trend in wild-type cells (Fig. 7), the data indicate that the improvement has a genetic basis. One possible explanation is that under continued cultivation, deleterious secondary mutations are selected against, and a purifying selection occurs.
The flow cytometric sorting approaches described here and elsewhere (Doan and Obbard 2012), which rely on isolation of clones only after repeated selection under competitive growth conditions, offer substantial and practical advantages over plating approaches. One major advantage is the requirement for competitive growth, which will eliminate clones with detrimental mutations affecting growth. In addition, the approach does not preselect for a particular genotype—in theory, any type of mutant that gives rise to a high lipid accumulating phenotype could be isolated by this method. This opens the door to using random mutagenesis not only to isolate useful phenotypes, but as a tool to identify the underlying genetic mechanisms. With high-throughput sequencing approaches, it is entirely feasible to sequence the genome and transcriptome of mutants for comparison with wild type to identify the locus of mutation. If only a few mutants generated by random mutagenesis are compared, then the presence of mutations throughout the genome may make identification of the responsible gene difficult. With larger numbers of mutants, the consistent presence of mutations in the same gene will highlight that gene as a good candidate which could be further tested by manipulation. The approach described in this report enabled isolation of large numbers of high performance mutants, in which multiple classes of mutations could be present. Characterization of multiple mutant classes could lead to the identification of the roles of various metabolic steps in the process of high TAG accumulation.
References
Aquatic Species Program Closeout Report (1998), National Renewable Energy Laboratory http://www.nrel.gov/docs/legosti/fy98/24190.pdf
Ball S, Marianne T, Diriek L, Fresnoy M, Delrue B, Decq A (1991) A Chlamydomonas reinhardtii low-starch mutant is defective for 3-phosphoglycerate activation and orthophosphate inhibition of ADP-glucose pyrophosphorylase. Planta 185:17–25
Cooper MS, Hardin WR, Petersen TW, Cattolico RA (2010) Visualizing “green oil” in live algal cells. J Biosci Bioeng 109:198–201
Delrue B, Fontaine T, Routier F, Decq A, Wieruszeski JM, van den Koornhuyse N, Maddelein ML, Fournet B, Ball S (1992) Waxy Chlamydomonas reinhardtii: monocellular algal mutants defective in amylose biosynthesis and granule-bound starch synthase activity accumulate a structurally modified amylopectin. J Bacteriol 174:3612–3620
Doan TY, Obbard JP (2012) Enhanced intracellular lipid in Nannochloropsis sp. via random mutagenesis and flow cytometric cell sorting. Algal Res 1:17–21
Doan TY, Obbard JP (2011) Enhanced lipid production in Nannochloropsis sp. using fluorescence-activated cell sorting. GCB Bioenergy 3:264–270
General Atomics (2012) Affordable algal-derived JP-8 surrogate. Project Final Report for Defense Advanced Research Projects Agency. (proprietary)
Hu Q, Sommerfeld M, Jarvis E, Ghirardi M, Posewitz M, Seibert M, Darzins A (2008) Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J 54:621–639
Huesemann MH, Hausmann TS, Bartha R, Aksoy M, Weissman JC, Benemann JR (2009) Biomass productivities in wild type and pigment mutant of Cyclotella sp (diatom). Appl Biochem Biotech 157:507–526
Montero MF, Aristizábal M, Reina GG (2011) Isolation of high-lipid content strains of the marine microalga Tetraselmis suecica for biodiesel production by flow cytometry and single-cell sorting. J Appl Phycol 23:1053–1057
Mutanda T, Ramesh D, Karthikeyan S, Kumari S, Anandraj A, Bux F (2011) Bioprospecting for hyper-lipid producing microalgal strains for sustainable biofuel production. Bioresource Tech 102:57–70
Pienkos PT, Laurens L, Ade A (2011) Making biofuel from microalgae. Amer Sci 99:474–481
Plumed MD, Villarejo A, de los Rios A, Garcia-Reina G, Ramazanov Z (1996) The CO2-concentrating mechanism in a starchless mutant of the green unicellular alga Chlorella pyrenoidosa. Planta 200:28–31
Wang ZT, Ullrich N, Joo S, Waffenschmidt S, Goodenough U (2009) Algal lipid bodies: stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardti Euk. Cell 8:1856–1868
Zabawinski C, Van den Koornhuyse N, D'Hulst C, Schlichting R, Giersch C, Delrue B, Lacroix JM, Preiss J, Ball S (2001) Starchless mutants of Chlamydomonas reinhardtii lack the small subunit of a heterotetrameric ADP-Glucose pyrophosphorylase. J Bacteriol 183:1069–1077
Acknowledgments
This work was supported by subcontract 4500024860 from General Atomics.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Manandhar-Shrestha, K., Hildebrand, M. Development of flow cytometric procedures for the efficient isolation of improved lipid accumulation mutants in a Chlorella sp. microalga. J Appl Phycol 25, 1643–1651 (2013). https://doi.org/10.1007/s10811-013-0021-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-013-0021-8