Abstract
Productivity of Indian mustard (Brassica juncea L. Czern. and Coss.) is markedly reduced by salt stress. To develop salt tolerance in this important oilseed crop is a need of the hour. This study, based on analysis of growth parameters and antioxidant profile of fourteen Indian mustard genotypes treated with 50, 100, 150 and 200 mM of sodium chloride, was performed to identify the salt-sensitive and salt-tolerant genotypes. Salinity stress inhibited biomass accumulation and reduced the protein and chlorophyll contents in a dose-dependent manner. The reduction was the highest in genotype Pusa Agrani and lowest in CS-54, depicting their contrasting sensitivity to salt stress. Salt treatments triggered a concentration-dependent overproduction of reactive-oxygen species and a concurrent upregulation of the expression of different antioxidants. Genotype CS-54 showed the least damage and maintained a high antioxidant level with almost each salt treatment, exhibiting its competence to withstand the damage provoked by salinity stress. Genotype Pusa Agrani, on the contrary, depicted a salt-sensitive nature by way of its very high lipid peroxidation and low intensity of antioxidants. These two genotypes were further investigated through gel-based proteomic approach, which resulted in the identification and quantification of 42 salinity-responsive proteins related to different metabolic modifications. Molecular processes, including photosynthesis, redox homeostasis, nitrogen metabolism, ATP synthesis, protein synthesis and degradation, signal transduction and respiratory pathways, have exhibited significant changes. The identified stress-responsive proteins could pave the way to develop salt tolerance in Indian mustard plant, thus sustaining its productivity under salinity.




Similar content being viewed by others
References
Abogadallah GM (2010) Antioxidative defense under salt stress. Plant Signal Behav 5:369–374
Aebi H (1984) Catalase in vitro. Meth Enzymol 105:121–126
Ahmad P, Hakeem KR, Kumar A, Ashraf M, Akram NA (2012) Salt-induced changes in photosynthetic activity and oxidative defense system of three cultivars of mustard (Brassica juncea L.). Afr J Biotechnol 11(11):2694–2703
Anderson ME (1985) Determination of glutathione and glutathione disulfide in biological samples. Meth Enzymol 113:548–555
Anjum NA, Ahmad I, Mohmood I, Pacheco M, Duarte AC, Pereira E, Umar S, Ahmad A, Khan NA, Iqbal M, Prasad MNV (2012) Modulation of glutathione and its related enzymes in plants’ responses to toxic metals and metalloids: a review. Environ Exp Bot 75:307–324
Anjum NA, Sofo A, Scopam A, Roychoudhury A, Gill SS, Iqbal M, Lukatkin AS, Pereira E, Duarte AC, Ahmad I (2014) Lipids and proteins—major targets of oxidative modifications in abiotic stressed plants. Environ Sci Pollut Res 22(6):4099–4121. doi:10.1007/s11356-014-3917-1
Aref IM, Khan PR, Khan S, El-Atta H, Ahmed AI, Iqbal M (2016) Modulation of antioxidant enzymes in Juniperus procera needles in relation to habitat environment and dieback incidence. Trees Struct Funct. doi:10.1007/s00468-016-1399-0
Arnon DI (1949) Copper enzymes in isolated chloroplasts. Polyphenoxidase in Beta vulgaris. Plant Physiol 24:1–15
Arshi A, Abdin MZ, Iqbal M (2004) Changes in biochemical status and growth performance of Senna (Cassia angustifolia Vahl.) grown under salt stress. Phytomorphology 54:109–124
Arshi A, Ahmad A, Are IM, Iqbal M (2012) Comparative studies on antioxidant enzyme action and ion accumulation in soybean cultivars under salinity stress. J Environ Biol 33:9–20
Ashraf M, Harris PJC (2004) Potential biochemical indicators of salinity tolerance in plants. Plant Sci 166:3–16
Bandehagh A, Salekdeh GH, Toorchi M, Mohammadi A, Komatsu S (2011) Comparative proteomic analysis of canola leaves under salinity stress. Proteomics 11:1965–1975
Bashir H, Qureshi MI, Ibrahim AM, Iqbal M (2015) Chloroplast and photosystems: impact of cadmium and iron deficiency. Photosynthetica 53:321–335
Beyer WF, Fridovich I (1987) Assaying for superoxide dismutase activity: some large consequences of minor changes in conditions. Anal Biochem 161:559–566
Boex-Fontvieille E, Daventure M, Jossier M, Hodges M, Zivy M, Tcherkez G (2014) Phosphorylation pattern of Rubisco activase in Arabidopsis leaves. Plant Biol 16:550–557
Bradford MM (1976) Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Busheva M, Tzonova I, Stoitchkova K, Andreeva A (2012) Heat-induced reorganization of the structure of photosystem II membranes: role of oxygen evolving complex. J Photochem Photobiol, B 117:214–221
Caruso G, Cavaliere C, Guarino C, Gubbiotti R, Foglia P, Lagana A (2008) Identification of changes in Triticum durum L. leaf proteome in response to salt stress by two-dimensional electrophoresis and MALDI-TOF mass spectrometry. Anal Bioanal Chem 391:381–390
Du CX, Fan HF, Guo SR, Tezuka T, Li J (2010) Proteomic analysis of cucumber seedling roots subjected to salt stress. Phytochemistry 71:1450–1459
Feller U, Anders I, Demirevska K (2008) Degradation of rubisco and other chloroplast proteins under abiotic stress. Gen Appl Plant Physiol 34:5–18
Foyer CH, Halliwell B (1976) The presence of glutathione and glutathione reductase in chloroplasts, A proposed role in ascorbic acid metabolism. Planta 133:21–25
Foyer CH, Noctor G (2009) Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antiox Redox Sign 11:861–905
Galmes J, Aranjuelo I, Medrano H et al (2013) Variation in Rubisco content and activity under variable climatic factors. Photosynth Res 117:73–90
Gapinska M, Skodowska M, Gabara B (2008) Effect of short- and long-term salinity on the activities of antioxidative enzymes and lipid peroxidation in tomato roots. Acta Physiol Plant 30:11–18
Gengmao Z, Yu Z, Xing S et al (2015) Salinity stress increases secondary metabolites and enzyme activity in safflower. Indust Crops Prod 64(1):175–181. doi:10.1016/j.indcrop.2014.10.058
Ghaffari A, Gharechahi J, Nakhoda B et al (2014) Physiology and proteome responses of two contrasting rice mutants and their wild type parent under salt stress conditions at the vegetative stage. J Plant Physiol 171:31–44
Guan Z, Chai T, Zhang Y et al (2009) Enhancement of Cd tolerance in transgenic tobacco plants overexpressing a Cd-induced catalase cDNA. Chemosphere 76(5):623–630
Hakeem KR, Chandana R, Ahmad P, Iqbal M, Ozturk M (2012a) Relevance of proteomic investigations in plant abiotic stress physiology. OMICS 16(11):621–635
Hakeem KR, Khan F, Chandna R, Siddiqui TO, Iqbal M (2012b) Genotypic variability among soybean genotypes under NaCl stress and proteome analysis of salt-tolerant genotype. Appl Biochem Biotechnol 168(8):2309–2329
Harinasut P, Poonsopa D, Roengmongkol K et al (2003) Salinity effects on antioxidant enzymes in mulberry cultivar. Sci Asia 29:109–113
Heath RL, Packer L (1968) Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophy 125:189–198
Hiscox JD, Israelstam GF (1979) A method for the extraction of chlorophyll from leaf tissue without maceration. Can J Bot 57:1332–1334
Hsieh TH, Li CH, Su RC et al (2010) A tomato bZIP transcription factor, SlAREB, is involved in water deficit and salt stress response. Planta 231:1459–1473
Huergo LF, Chandra G, Merrick M (2013) PII signal transduction proteins: nitrogen regulation and beyond. FEMS Microbiol Rev 37:251–283
Iqbal M, Ahmad A, Ansari MKA et al (2015) Improving the phytoextraction capacity of plants to scavenge metal(loid)-contaminated sites. Environ Rev 23:44–65
Isaacson T, Damasceno CM, Saravanan RS et al (2006) Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nat Protoc 1(2):769–774
Jeong MJ, Park SC, Byun MO (2001) Improvement of salt tolerance in transgenic potato plants by glyceraldehyde-3 phosphate dehydrogenase gene transfer. Mol Cells 12:185–189
Ji H, Pardo JM, Batelli G et al (2013) The salt overly sensitive (SOS) pathway: established and emerging roles. Mol Plant 6(2):275–286
Jiang Y, Yang B, Harris NS et al (2007) Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J Exp Bot 58:3591–3607
Kim Y, Arihara J, Nakayama T, Nakayama N (2004) Antioxidative responses and their relation to salt tolerance in Echinochloa oryzicola Vasing and Steraia virdis (L.) Beauv. Plant Growth Regul 44:87–92
Kim DW, Rakwal R, Agrawal GK et al (2005) A hydroponic rice seedling culture model system for investigating proteome of salt stress in rice leaf. Electrophor 26:4521–4539
Kong-Ngern K, Daduang S, Wongkham CH et al (2005) Protein profiles in response to salt stress in leaf sheaths of rice seedlings. Sci Asia 31:403–408
Kukreja S, Nandval AS, Kumar N et al (2005) Plant water status, H2O2 scavenging enzymes, ethylene evolution and membrane integrity of Cicer arietinum roots as affected by salinity. Biol Plant 49:305–308
Kulik A, Mazur AA, Bucholc M et al (2004) Antioxidant responses of shoots and roots of lentil to NaCl salinity stress. Plant Growth Regul 42:69–77
Kulik A, Wawer I, Krzywinska E et al (2011) SnRK2 protein kinases-key regulators of plant response to abiotic stresses. OMICS 15:859–872
Law MY, Charles SA, Halliwell B (1983) Glutathione and ascorbic acid in spinach chloroplasts. Biol Biochem J 210:899–903
Lei P, Xu Z, Liang J, Luo X, Zhang Y, Feng X, Xu H (2016) Poly (γ-glutamic acid) enhanced tolerance to salt stress by promoting proline accumulation in Brassica napus L. Plant Growth Regul 78(2):233–241
Li C, Lv J, Zhao X et al (2010) TaCHP: a wheat zinc finger protein gene down-regulated by abscisic acid and salinity stress plays a positive role in stress tolerance. Plant Physiol 154:211–221
Lu S, Li T, Jiang J (2010) Effects of salinity on sucrose metabolism during tomato fruit development. Afr J Biotech 9(6):842–849
Lyzenga WJ, Stone SL (2012) Abiotic stress tolerance mediated by protein ubiquitination. J Exp Bot 63(2):599–616
Manaa A, Mimouni H, Wasti S, Gharbi E, Aschi-Smiti S, Faurobert M, Ahmed HB (2013) Comparative proteomic analysis of tomato (Solanum lycopersicum) leaves under salinity stress. Plant Omics J 6(4):268–277
Medeiros CD, Oliveira MT, Rivas R et al (2012) Gas exchange, growth, and antioxidant activity in sugarcane under biological nitrogen fixation. Photosynthetica 50:519–528
Meskiene I, Baudouin E, Schweighofer A et al (2003) Stress-induced protein phosphatase 2C Is a negative regulator of a mitogen-activated protein kinase. J Biol Chem 278(21):18945–18952
Miranda RS, Gomes-Filho E, Prisco JT, Alvariz-Pizzaro JC (2016) Ammonium improves tolerance to salinity stress in Sorghum bicolor plants. Plant Growth Regul 78(1):121–131
Munns R, James RA (2003) Screening methods for salinity tolerance: a case study with tetraploid wheat. Plant Soil 253:201–218
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Ann Rev Plant Biol 59:651–681
Murata N, Takahashi S, Nishiyama Y, Allakhverdiev SI (2007) Photoinhibition of photosystem II under environmental stress. Biochim Biophys Acta 1767:414–421
Nakano Y, Asada K (1981) Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol 22:867–880
Nazar R, Iqbal N, Syeed S et al (2011) Salicylic acid alleviates decreases in photosynthesis under salt stress by enhancing nitrogen and sulfur assimilation and antioxidant metabolism differentially in two mungbean cultivars. J Plant Physiol 168:807–815
O’Farrel PH (1975) High resolution two-dimensional electrophoresis of proteins. J Biol Chem 250:4007–4021
Parihar R, Singh S, Singh R et al (2015) Effect of salinity stress on plants and its tolerance strategies: a review. Environ Sci Pollut Res 22:4056–4075
Parker R, Flowers TJ, Moore AL et al (2006) An accurate and reproducible method for proteome profiling of the effects of salt stress in the rice leaf. J Exp Bot 57:1109–1118
Paszkowski U, Kroken S, Roux C, Briggs S (2002) Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 99:13324–13329
Puig J, Meynard D, Khong GN et al (2013) Analysis of the expression of the AGL17-like clade of MADS-box transcription factors in rice. Gene Express Patter 13:160–170
Qadir M, Schubert S (2002) Degradation processes and nutrient constraints in sodic soils. Land Degrad Develop 13(4):275–294
R Core Team (2015) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.r-project.org/
Rais L, Masood A, Inam A, Khan N (2013) Sulfur and nitrogen co-ordinately improve photosynthetic efficiency, growth and proline accumulation in two cultivars of mustard under salt stress. J Plant Biochem Physiol 1:101
Rao MV (1992) Cellular detoxifying mechanisms determine age dependent injury in tropical plants exposed to SO2. J Plant Physiol 140:733–740
Rowley ER, Mockler TC (2011) Plant abiotic stress: insights from the genomics era. In: Shankar A, Venkateswarlu B (eds) Abiotic stress response in plants—physiological, biochemical and genetic perspectives. InTech Open, Rijeka, Croatia, pp 221–268
Sahi C, Singh A, Blumwald E, Grover A (2006) Beyond osmolytes and transporters: novel plant salt stress tolerance-related genes from transcriptional profiling data. Physiol Plant 127:1–9
Santos CV (2004) Regulation of chlorophyll biosynthesis and degradation by salt stress in sunflower leaves. Sci Hort 103:93–99
Shah SH (2007) Effects of salt stress on mustard as affected by gibberellic acid application. Gen App Plant Physiol 33(1–2):9–106
Sharma P, Jha AB, Dubey RS et al (2012) Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J Bot 2012:1–26
Sharma P, Sardana V, Banga SS (2013) Salt tolerance of Indian mustard (Brassica juncea) at germination and early seedling growth. Environ Exp Biol 11:39–46
Shekhawat GS, Verma K (2010) Heme oxygenase (HO): an overlooked enzyme of plant metabolism and defence. J Exp Bot 61(9):2255–2270
Sumithra K, Jutur PP, Carmel BD, Reddy AR (2006) Salinity-induced changes in two cultivars of Vigna radiata: responses of antioxidative and proline metabolism. Plant Growth Regul 50:11–22
Suorsa M, Aro EM (2007) Expression, assembly and auxiliary functions of photosystem II oxygen-evolving proteins in higher plants. Photosynth Res 93:89–100
Takahashi S, Badger MR (2011) Photoprotection in plants: a new light on photosystem II damage. Trends Plant Sci 16(1):53–60
Uhrig GR, Kenneth KS, Greg BG et al (2009) PII in higher plants: a modern role for an ancient protein. Trend Plant Sci 14(9):505–511
Umezawa T, Yoshida R, Maruyama K et al (2004) SRK2C a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana. PNAS 101(49):17306–17311
Venkatesh J, Upadhyaya CP, Yu JW, Hemavathi A, Kim DH, Strasser RJ, Park SW (2012) Chlorophyll a fluorescence transient analysis of transgenic potato overexpressing d-galacturonic acid reductase gene for salinity stress tolerance. Hort Environ Biotechnol 53:320–328
Wang W, Vinocur B, Altman A (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218:1–14
Wang X, Wei Z, Liu D et al (2011) Effects of NaCl and silicon on activities of antioxidative enzymes in roots, shoots and leaves of alfalfa. Afr J Biotechnol 10(4):545–549
Yamamoto M, Nasrallah JB (2009) In planta assessment of the role of thioredoxin h proteins in the regulation of S-locus receptor kinase signaling in transgenic Arabidopsis. Plant Physiol 163:1387–1395
Yasar F, Ellialtioglu S, Yildiz K (2008) Effect of salt stress on antioxidant defense systems, lipid peroxidation and chlorophyll content in green bean. Russ J Plant Physiol 55(6):782–786
Yousuf PY, Hakeem KR, Chandna R, Ahmad P (2012) Role of glutathione reductase in plant abiotic stress. In: Ahmad P, Prasad MNV (eds) Abiotic stress responses in plants. Springer, New York, pp 149–158
Yousuf PY, Ahmad A, Ganie AH, Iqbal M (2016a) Salt stress-induced modulations in the shoot proteome of Brassica juncea genotypes. Environ Sci Pollut Res 23(3):2391–2401. doi:10.1007/s11356-015-5441-3
Yousuf PY, Ahmad A, Aref IM, Ozturk M, Hemant Ganie AH, Iqbal M (2016b) Salt-stress-responsive chloroplast proteins in Brassica juncea genotypes with contrasting salt tolerance and their quantitative PCR analysis. Protoplasma. doi:10.1007/s00709-015-0917-z
Zhang C, Liu J, Zhang Y, Cai X, Gong P, Zhang J, Wang T, Li H, Ye Z (2011) Overexpression of SlGMEs leads to ascorbate accumulation with enhanced oxidative stress, cold, and salt tolerance in tomato. Plant Cell Rep 30:389–398
Acknowledgments
The first author is grateful to Hamdard National Foundation (HNF) New Delhi, India for granting a fellowship. The second author (A Ahmad) is now working at the Department of Botany, Aligarh Muslim University, Aligarh 202002, India.
Author contributions
P. Y. Yousuf conducted experimental research with the help of A. Ahmad and A. H. Ganie. Statistical analysis was done by O Sareer and V Krishnapriya. Data interpretation and MS preparation were done by M. Iqbal, A. Ahmad and I. M. Aref.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors have no conflict of interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10725_2016_182_MOESM1_ESM.jpg
Fig. S1: Plants of salt-tolerant genotype (CS-54) grown under (a) control and (b) 200 mM NaCl treatment and salt-sensitive genotype Pusa Agrani grown under (c) control and (d) 200 mM NaCl treatment, at the time of sampling (JPEG 244 kb)
10725_2016_182_MOESM2_ESM.jpg
Fig. S2: (a) Clustering and (b) PCA analysis based on biomass of 14 different genotypes of Indian mustard grown under salt stress. Correlation matrix was used for PCA. Clustering was done using the Ward’s method of Squared Euclidean distance matrix. PCA and clustering of biomass clearly differentiate V2 and V5. Arrows for all traits are directed towards V5 indicating that it is indeed a tolerant variety, whereas V2 presents an opposite case indicating its salt-sensitive nature. Cluster dendrogram also reflects similar results (JPEG 1077 kb)
10725_2016_182_MOESM3_ESM.jpg
Fig. S3: PCA analysis of 14 different genotypes of Indian mustard under different salt treatments (a) T0 (b) T1 (c) T2 (d) T3 (e) T4, based on physiological and biochemical parameters. Percentage values given in brackets in the axis labels denote R2 values (JPEG 1586 kb)
10725_2016_182_MOESM4_ESM.jpg
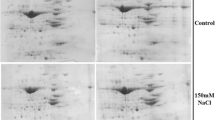
Fig. S4: Spot images of five representative proteins differentially-expressed during different salt treatments in salt-sensitive (Pusa Agrani) and salt-tolerant (CS-54) genotypes of Indian mustard (JPEG 189 kb)
Rights and permissions
About this article
Cite this article
Yousuf, P.Y., Ahmad, A., Ganie, A.H. et al. Antioxidant response and proteomic modulations in Indian mustard grown under salt stress. Plant Growth Regul 81, 31–50 (2017). https://doi.org/10.1007/s10725-016-0182-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-016-0182-y