Abstract
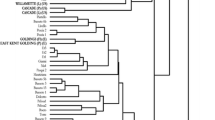
Microsatellite variation at the nuclear and chloroplast genomes was evaluated for wild European and wild American hops, in order to assess the genetic diversity and origin of cultivated hops. Seven nuclear loci and 32 chloroplast loci were used in the analysis of 182 hop accessions including wild European (68), wild American (48), and cultivars (66). A total of 116 alleles were identified using 7 nuclear microsatellites showing different averages of polymorphism and distribution in the wild American and European accessions and cultivars. Two main groups were established as revealed by several statistical analyses; one including European wild accessions and cultivars and a second group consisting of American wild accessions. Three polymorphic chloroplast microsatellite loci were detected, six alleles were scored which defined a total of five haplotypes that were exclusive or presented different distribution between American and European wild accessions. A major influence of the wild European haplotypes was detected among hop cultivars. To the best of our knowledge, this is the first work reporting the use of chloroplast microsatellites in hops.



Similar content being viewed by others
References
Arroyo-García R, Lefort F, De Andrés MT, Ibañez J, Borrego J, Jouve N, Cabello F, Martínez-Zapater JM (2002) Chloroplast microsatellite polymorphism in Vitis species. Genome 45:1142–1149. doi:10.1139/G02-087
Arroyo-García R, Ruiz-García L, Bolling L, Ocete R, López MA, Arnold C, Ergul A, Söylemezolu G, Uzun HL, Cabello F, Ibáñez J, Aradhya MK, Atanassov A, Atanassov I, Balint S, Cenis JL, Costantini L, Gorislavets S, Grando MS, Klein BY, Mcgovern PE, Merdinoglu D, Pejic I, Pelsy F, Primikirios N, Risovannaya V, Roubelakis-Angelakis KA, Snoussi H, Sotiri P, Tamhankar S, This P, Troshin L, Malpica JM, Lefort F, Martinez-Zapater JM (2006) Multiple origins of cultivated grapevine (Vitis vinifera L. ssp. sativa) based on chloroplast DNA polymorphisms. Mol Ecol 15:3707–3714. doi:10.1111/j.1365-294X.2006.03049.x
Bandelt HJ, Forster P, Sykes BC, Richards MB (1995) Mitochondrial portraits of human populations. Genetics 141:743–753
Bowers J, Boursiquot JM, This P, Chu K, Johansson H, Meredith C (1999) Historical genetics: the parentage of Chardonnay, Gamay, and other wine grapes of Northeastern France. Science 285:1562–1565. doi:10.1126/science.285.5433.1562
Brady JL, Scott NS, Thomas MR (1996) DNA typing of hops (Humulus lupulus L.) through application of RAPD microsatellite marker sequences converted to sequence tagged sites. Euphytica 91:277–284
Cerenak C, Jakše J, Javornik B (2004) Identification and differentiation of hop varieties using simple sequence repeat markers. J Am Brew Chem 62:1–7
Chung SM, Staub JE (2003) The development and evaluation of consensus chloroplast primer pairs that posses highly variable sequence regions in diverse array of plant taxa. Theor Appl Genet 107:757–767
Darby P (1999) New selection criteria in hop breeding. In: Seigner E (ed) International hop growers convention. Proceedings of the scientific commission. Pulawy, pp 3–6
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. doi:10.1111/j.1365-294X.2005.02553.x
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinf Online 1:47–50
Hadonou AM, Walden R, Darby P (2004) Isolation and characterization of polymorphic microsatellites for assessment of genetic variation in hops (Humulus lupulus L.). Mol Ecol Notes 4:280–282. doi:10.1111/j.1471-8286.2004.00641.x
Hammer Ø, Harper DAT, Ryan PD (2006) PAST—PAlaeontological STatistics, ver 1.54. Distributed by the author. http://folk.uio.no/ohammer/past/download.html
Jakše J, Kindlhofer K, Javornik B (2001) Assessment of genetic variation and differentiation in hop (Humulus lupulus L.) genotypes by microsatellite and AFLP markers. Genome 44:773–782. doi:10.1139/gen-44-5-773
Jakše J, Bandelj D, Javornik B (2002) Eleven new microsatellites for hops (Humulus lupulus L.). Mol Ecol Notes 2:544–546. doi:10.1046/j.1471-8286.2002.00309.x
Jakše J, Satovic Z, Javornik B (2004) Microsatellite variability among wild and cultivated hops (Humulus lupulus L.). Genome 47:889–899. doi:10.1139/G04-054
Jakše J, Luthar Z, Javornik J (2008) New polymorphic dinucleotide and trinucleotide microsatellite loci for hop Humulus lupulus L. Mol Ecol Res 8:769–772. doi:10.1111/j.1471-8286.2007.02053.x
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Levinson G, Gutman GA (1987) Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol 4:203–221
Moir M (2000) Hop—a millennium review. J Am Soc Brew Chem 58:131–146. doi:10.1094/ASBCJ-58-0131
Murakami A, Darby P, Javornik B, Pais MSS, Seigner E, Luzt A, Svoboda P (2006a) Molecular phylogeny of wild hops, Humulus lupulus L. Heredity 97:66–74. doi:10.1038/sj.hdy.6800839
Murakami A, Darby P, Javornik B, Pais MSS, Seigner E, Luzt A, Svoboda P (2006b) Microsatellite DNA analysis of wild hops, Humulus lupulus L. Genet Resour Crop Evol 53:1553–1562. doi:10.1007/s10722-005-7765-1
Neve RA (1991) Hops. Chapman and Hall, London, pp 1–78
Patzak J (2002) Characterization of Czech hop (Humulus lupulus L.) genotypes by molecular methods. Plant Prod 48:343–350
Patzak J, Vrba L, Matousek J (2007) New SSR molecular markers for assessment of genetic diversity and DNA fingerprinting in hop (Humulus lupulus L.). Genome 50:15–25. doi:10.1139/G06-128
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295. doi:10.1111/j.1471-8286.2007.01787.x
Peredo EL, Arroyo-Garcia R, Martinez-Zapater JM, Revilla MA (2005) Evaluation of microsatellite detection using autoradiography and capillary electrophoresis in hops. J Am Brew Chem 63:57–62. doi:10.1094/ASBCJ-63-0057
Pillay M, Kenny ST (1994) Chloroplast DNA differences between cultivated hop, Humulus lupulus and the related species H. japonicus. Theor Appl Genet 89:373–378
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotypes data. Genetics 155:945–959
Provan J, Powell W, Hollingsworth PM (2001) Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends Ecol Evol 16:142–147. doi:10.1046/j.1365-294X.2000.105316.x
Seefelder S, Ehrmaier H, Schweizer G, Seigner E (2000) Genetic diversity and phylogenetic relationships among accessions of hop, Humulus lupulus, as determined by amplified fragment length polymorphism fingerprinting compared to pedigree data. Plant Breed 119:257–263. doi:10.1111/j.1439-0523.2000.tb01637.x
Siushih C (1998) Cannaboideae. In: Siushih C, Wu C (eds) Fl Republ Popularis Sin, vol 23, pp 220–224
Small E (1978) A numerical and nomenclatural analysis of morpho-geographic taxa of Humulus lupulus. Sys Bot 3:37–76
Stajner N, Jakše J, Kozjak P, Javornik B (2005) The isolation and characterization of microsatellite in hop (Humulus lupulus L.). Plant Sci 168:213–221. doi:10.1016/j.plantsci.2004.07.031
Stajner N, Satovic Z, Cerenak A, Javornik B (2008) Genetic structure and differentiation in hop (Humulus lupulus L.) as inferred from microsatellite. Euphytica 161:301–311. doi:10.1007/s10681-007-9429-z
Tautz D (1989) Hypervariability of simple sequences as general source for polymorphic markers. Nucleic Acids Res 17:6463–6471
Townsend MS, Henning JA (2009) AFLP discrimination of native North American and cultivated hop. Crop Sci 42:600–607
Weising K, Gardner RC (1999) A set of conserved PCR primers for the analysis of simple sequence repeat polymorphism in chloroplast genomes of dicotyledonous angiosperms. Genome 42:9–19
Acknowledgments
This research was supported by funds from the Spanish Ministry of Science and Technology and FEDER (project MCT-02-AGL-02472) and the project CGL2005-06821-C02-01. E.L.P. is in receipt of an F.P.U. grant. The plant material was provided by the USDA-ARS National Clonal Germplasm Repository (Corvallis, OR, USA) supported by CRIS 5358-21000-033-00D and by Branka Javornik from the University of Ljubljana and Slovene Institute of Hop Research and Brewing (Zalec, Slovenia). Thanks are given to the people in the Departamento de Biotecnología (INIA) where part of this research was carried out. Thanks also to Leo Blanco and Fernando Rodríguez for their statistical assistance. The authors want to thank Dr. Jernej Jakse and Fernando Rodríguez for their critical pre-view and comments which helped to improve this paper.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Peredo, E.L., Ángeles Revilla, M., Reed, B.M. et al. The influence of European and American wild germplasm in hop (Humulus lupulus L.) cultivars. Genet Resour Crop Evol 57, 575–586 (2010). https://doi.org/10.1007/s10722-009-9495-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-009-9495-2