Abstract
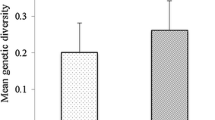
Information regarding the amount of genetic diversity is necessary to enhance the effectiveness of breeding programs and germplasm conservation efforts. Genetic variation between 21 switchgrass genotypes randomly selected from two lowland (‘Alamo’ and ‘Kanlow’) and one upland (‘Summer’) synthetic cultivars were estimated using restriction fragment length polymorphism (RFLP) markers. Comparison of 85 RFLP loci revealed 92% polymorphism between at least two genotypes from the upland and lowland ecotypes. Within ecotypes, the upland genotypes showed higher polymorphism than lowland genotypes (64% vs. 56%). ‘Kanlow’ had a lower percent of polymorphic loci than ‘Alamo’ (52% vs. 60%). Jaccard distances revealed higher genetic diversity between upland and lowland ecotypes than between genotypes within each ecotype. Hierarchical cluster analysis using Ward's minimum variance grouped the genotypes into two major clusters, one representing the upland group and the other the lowland group. Phylogenetic analysis of chloroplast non-coding region trnL (UAA) intron sequences from 34 switchgrass accessions (6 upland cultivars, 2 lowland cultivars, and 26 accessions of unknown affiliation) produced a neighbor-joining dendrogram comprised of two major clusters with 99% bootstrap support. All accessions grouped in the same cluster with the lowland cultivars (‘Alamo’ and ‘Kanlow’) had a deletion of 49 nucleotides. Phenotypic identification of greenhouse-grown plants showed that all accessions with the deletion are of the lowland type. The deletion in trnL (UAA) sequences appears to be specific to lowland accessions and should be useful as a DNA marker for the classification of upland and lowland germplasm.
Similar content being viewed by others
Abbreviations
- RFLP:
-
restriction fragment length polymorphism
- trnL (UAA):
-
tRNALeucine
References
Alderson J. and Sharp W.C. (1995). Grass Varieties in the United States. CRC Lewis Publishers, Boca Raton, FL
Bhattacharjee R., Bramel P., Hash C.T., Kolesnikova-Allen M. and Khairwal I. (2002). Assessment of genetic diversity within and between pearl millet landraces. Theor. Appl. Genet. 105: 666–673
Burton G.W. (1942). A cytological study of some species in the tribe Paniceae. Am. J. Bot. 29: 355–359
Causse M., Fulton T.M., Cho Y.G., Ahn S.N., Chunwongse J., Wu K., Xiao J., Yu Z., Ronald P.C., Harrington S.B., Second G.A., McCouch S.R. and Tanksley S.D. (1994). Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics 138: 1251–1274
Church G.L. (1940). Cytotaxonomic studies in the gramineae Spartina, Andropogon, and Panicum. Am. J. Bot. 27: 263–271
Clegg M.T., Cummings M.P. and Durbin M.L. (1997). The evolution of plant nucleargenes. Proc. Nat. Acad. Sci. USA 94: 7791–7798
Falouss A. (1989). Distributional Properties of Jaccard's Index of Similarity. Thesis (M.S.), University of Georgia, Athens, GA
Feinberg A.P. and Vogelstein B. (1983). A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132: 6–13
Felsenstein J. (1985). Confidence limits on phylogenies: an approach using bootstrap. Evolution 39: 783–791
Gauthier P., Gouesnard B., Dallard J., Redaelli R., Rebourg C., Charcosset A. and Boyat A. (2002). RFLP diversity and relationships among traditional European maize populations. Theor. Appl. Genet. 105: 91–99
Gielly L., Yuan Y.M., Kupfer P. and Taberlet P. (1996). Phylogenetic use of noncoding regions in the genus Gentiana L.: chloroplast trnL (UAA) intron versus nuclear ribosomal internal transcribed spacer sequences. Mol. Phylogenet. Evol. 5: 460–466
Gunter L.E., Tuskan G.A. and Wullschleger S.D. (1996). Diversity among populations of switchgrass based on RAPD markers. Crop Sci. 36: 1017–1022
Hein J. (1990). Unified approach to alignments and phylogenies. Methods Enzymol. 183: 625–645
Henry D.S. and Taylor T.H. (1989). Registration of KY1625 switchgrass germplasm. Crop Sci. 29: 1096
Hitchcock A.S. (1971). Manual of Grasses of the United States. Vol. II. Dover Publications Inc., New York.
Hopkins A.A. and Taliaferro C.M. (1995). A comparison of selection strategies in switchgrass. Proc. Am. Forage Grass Council 4: 190–192
Hopkins A.A., Taliaferro C.M., Murphy C.D. and Christian D. (1996). Chromosome number and nuclear DNA content of several Switchgrass population. Crop Sci. 36: 1192–1195
Huang S., Su X., Haselkorn R. and Gornicki P. (2003). Evolution of switchgrass (Panicum virgatum L.) based on sequences of the nuclear gene encoding plastid acetyl-CoA carboxylase. Plant Sci. 164: 43–49
Hultquist S.J., Vogel K.P., Lee D.J., Arumuganathan K. and Kaeppler S. (1996). Chloroplast DNA and nuclear DNA content variations among cultivars of switchgrass, Panicum virgatum L. Crop Sci. 36: 1049–1052
Jung G.A., Shaffer J.A., Stout W.L. and Panciera M.J. (1990). Warm season grass diversity in yield, plant morphology, and nitrogen concentration and removal in northeastern USA. Agron. J. 82: 21–26
Karp A., Seberg O. and Buiatti M. (1996). Molecular techniques in the assessment of botanical diversity. Ann. Bot. 78: 143–149
Kidwell K.K. and Osborn T.C. (1992). Simple plant DNA isolation procedures. In: Beckmann, J.S. and Osborn, T.C. (eds) Plant Genomes: Methods for Genetic and Physical Mapping, pp 1–13. Kluwer Academic Publishing, Netherlands
Lefort P. and Douglas G.C. (1999). An efficient micro method of DNA isolation from mature leaves of four hardwood tree species Acer, Fraxinus, Prunus and Quercus. Ann. Forest Sci. 56: 259–263
Martinez-Reyna J.M., Vogel K.P., Caha C. and Lee D.J. (2001). Meiotic stability, chloroplast DNA polymorphisms, and morphological traits of Upland Lowland switchgrass reciprocal hybrids. Crop Sci. 41: 1579–1583
Martinez-Reyna J.M. and Vogel K.P. (2002). Incompatibility systems in switchgrass. Crop Sci. 42: 1800–1805
Massa A.N., Larson S.R., Jensen K.B. and Hole D.J. (2001). AFLP variation in Bromus section Ceratochloa germplasm of Patagonia. Crop Sci. 41: 1609–1616
Missaoui A.M. (2003). Molecular Phylogenetic Analysis, Genetic Mapping and Improvement of Switchgrass (Panicum virgatum L.) for Bioenergy and Bioremediation to Excess Phosphorus in the Soil. Ph.D. dissertation, The University of Georgia, Athens, GA
Murray M.G. and Thompson W.F. (1982). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8: 4321–4325
Nei M. and Li W. (1979). Mathematical model for studying the genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76: 427–434
Newell L.C. (1936). Annual report, grass improvement investigations, USDA and the Nebraska AES. Lincoln, NE
Nielson E.L. (1944). Analysis of variation in Panicum virgatum. J. Agric. Res. 69: 327–353
Porter C.L. (1966). An analysis of variation between upland and lowland switchgrass Panicum virgatum L. in central Oklahoma. Ecology 47: 980–992
Redfearn D.D., Moore K.J., Vogel K.P, Waller S.S. and Mitchell R.B. (1999). Fiber digestion dynamics of sward components within switchgrass populations. Crop Sci. 39: 784–789
Reiter R.S., Williams J.G.K, Feldmann K.J., Rafalski J.A., Tingey S.C. and Scolnik P.A. (1992). Global and local genome mapping in Arabidopsis thaliana by using recombinant inbred lines and random amplified polymorphic DNAs. Proc. Natl. Acad. Sci. USA. 89: 1477–1488
Saitou N. and Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425
Sanderson M.A., Reed R.L., McLaughlin S.B., Wullschleger S.D., Conger B.V., Parrish D.J., Wolf D.D., Taliaferro C., Hopkins A.A., Osumpaugh W.R., Hussey M.A., Read J.C. and Tischler C.R. (1996). Switchgrass as a sustainable bioenergy source. Bioresource Technol. 56: 83–93
SAS Institute Inc. 1999. SAS online doc, Version 8. SAS Institute Inc., Cary, NC
Southern E.M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98: 503–517
Sun Y., Skinner D.Z., Liang G.H. and Hulbert S.H. 1994. Phylogenetic analysis of sorghum and related taxa using internal transcribed spacers of nuclear ribosomal DNA. Theor. Appl. Genet. 89: 26–32
Swofford D.L. 2000. PAUP* 4.0 Beta Version: Phylogenetic analysis using Parsimony and other methods (software). Sinauer Associates, Sunderland, MA
Taberlet P., Gielly L., Pautou G. and Bouvet J. 1991. Universal primers for amplification of three noncoding regions of chloroplast DNA. Plant Mol. Biol. 17: 1105–1109
Talbert L.E., Timothy D.H., Burns J.C., Rawlings J.O. and Moll R.H. 1983. Estimates of genetic parameters in switchgrass. Crop Sci. 23: 725–728
Taliaferro C.M. and Hopkins A.A. 1996. Breeding characteristics and improvement potential of switchgrass. In: Cundiff, J.S. (eds) Proceedings of the third liquid fuel conference, Nashville, TN. 15–17 Sept. 1996, pp 2–9. ASAESt. Joseph, MI
Tamura K. and Nei M. 1993. Model selection in the estimation of the number of nucleotide substitutions. Mol. Biol. Evol. 10: 512–526
Thorman C.E. and Osborn T.C. 1992. Use of RAPD and RFLP markers for germplasm evaluation. In: (eds) Application of RAPD technology to plant breeding. Proceedings of the Joint Plant Breeding Symposia Series, 1992, pp 9–11. Minneapolis, Minnesota
Vogel K.P., Dewald C.L., Gorz H.J. and Haskins F.A. 1985. Development of switchgrass, indiangrass, and eastern gamagrass: Current status and future. In: (eds) Symposium on range plant improvement in western North America: Current status and future. Salt Lake City, UT, Soc Range Management, Denver, CO. pp 51–62.
Vogel K.P., Hopkins A.A., Moore K.J., Johnson K.D. and Carlson I.T. 1996. Registration of ‘Shawnee’ switchgrass. Crop Sci. 36: 1713
Ward J.H. (1963). Hierarchical grouping to optimize an objective function. Am. Stat. Assoc. J. 56: 236–244
Williams W.M., Ansari H.A., Ellison N.W. and Hussain S.W. 2001. Evidence of three subspecies in Trifolium nigrescens Viv. Ann. Bot. 87: 683–691
Wolda H. (1981). Similarity indices, sample size and diversity. Ecologia 50: 296–302
Zhang Q., Saghai-Maroof M.A., Lu T.Y. and Shen B.Z. 1992. Genetic diversity and differentiation of indica and japonica rice detected by RFLP analysis. Theor. Appl. Genet. 83: 495–499
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Missaoui, A.M., Paterson, A.H. & Bouton, J.H. Molecular Markers for the Classification of Switchgrass (Panicum virgatum L.) Germplasm and to Assess Genetic Diversity in Three Synthetic Switchgrass Populations. Genet Resour Crop Evol 53, 1291–1302 (2006). https://doi.org/10.1007/s10722-005-3878-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-005-3878-9