Abstract
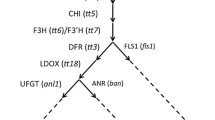
Two genes (DFR-A and ANS) encoding dihydroflavonol 4-reductase (DFR) and anthocyanidin synthase (ANS) enzymes in the anthocyanin biosynthesis pathway, respectively, are complementarily involved in anthocyanin production in onion (Allium cepa L.). Eleven inactive DFR-A alleles have been reported, with only a single inactive ANS allele previously identified. Two additional inactive ANS alleles are reported in this study. A mutant ANS allele containing a 4-bp insertion at the end of exon1 was identified from yellow bulbs of the F2 population in which the DFR-A genotype was homozygous for an active allele. The 4-bp insertion caused a frame-shift mutation and resulted in creation of a premature stop codon at the start of exon2. This mutant ANS allele was designated ANS PS allele. RT-PCR results showed that transcripts of the ANS PS allele were almost undetectable in yellow F2 bulbs, implying the involvement of nonsense-mediated mRNA decay. A cleaved amplified polymorphic sequence marker was developed for detection of the ANS PS allele. Another inactive ANS allele was identified from the light-red F1 populations showing complementation between DFR-A and ANS genes. A critical amino acid change of the strictly conserved serine residue into leucine was found in this mutant allele designated ANS S188L. In addition, seven variants of active ANS alleles were identified from diverse onion germplasm. A stepwise process consisting of PCR amplification and sequencing of PCR products was devised to identify three inactive (ANS PS, ANS S188L, and ANS G229R), one leaky (ANS P), and two active ANS alleles (ANS L and ANS h1).






Similar content being viewed by others
References
Abrahams S, Lee E, Walker AR, Tanner GJ, Larkin PJ, Ashton AR (2003) The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. Plant J 35:624–636
Antonescu C, Antonescu V, Sultana R, Quackenbush J (2010) Using the DFCI gene index databases for biological discovery. Curr Protoc Bioinform. doi:10.1002/0471250953.bi0106s29
Brewster JL (1994) Onions and other vegetable alliums. CAB International, Wallingford, pp 1–26
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552
Chang Y, Imam JS, Wilkinson MF (2007) The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 76:51–74
Clarke AE, Jones HA, Little TM (1944) Inheritance of bulb color in the onion. Genetics 29:569–575
Clere N, Faure S, Martinez MC, Andriantsitohaina R (2011) Anticancer properties of flavonoids: roles in various stages of carcinogenesis. Cardiovasc Hematol Agents Med Chem 9:62–77
Cook NC, Samman S (1996) Flavonoids—chemistry, metabolism, cardioprotective effects, and dietary sources. Nutr Biochem 7:66–76
Cushnie TP, Lamb AJ (2011) Recent advances in understanding the antibacterial properties of flavonoids. Int J Antimicrob Agents 38:99–107
Czemmel S, Heppel SC, Bogs J (2012) R2R3 MYB transcription factors: key regulators of the flavonoid biosynthetic pathway in grapevine. Protoplasma 249:S109–S118
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Duangjit J, Welsh K, Wise ML, Bohanec B, Havey MJ (2014) Genetic analyses of anthocyanin concentrations and intensity of red bulb color among segregating haploid progenies of onion. Mol Breed 34:75–85
El-Shafie MW, Davis GN (1967) Inheritance of bulb color in the onion (Allium cepa L.). Hilgardia 38:607–622
Ferrer JL, Austin MB, Stewart C Jr, Noel JP (2008) Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol Biochem 46:356–370
Fini A, Brunetti C, Di Ferdinando M, Ferrini F, Tattini M (2011) Stress-induced flavonoid biosynthesis and the antioxidant machinery of plants. Plant Signal Behav 6:709–711
Fossen T, Andersen OM, Ovstedal DO, Pedersen AT, Raknes A (1996) Characteristic anthocyanin pattern from onions and other Allium spp. J Food Sci 61:703–706
Goodrich J, Carpenter R, Coen ES (1992) A common gene regulates pigmentation pattern in diverse plant species. Cell 68:955–964
Griffiths G, Trueman L, Crowther T, Thomas B, Smith B (2002) Onions—a global benefit to health. Phytother Res 16:603–615
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Window 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hentze MW, Kulozik AE (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell 96:307–310
Herman-Antosiewicz A, Singh SV (2004) Signal transduction pathways leading to cell cycle arrest and apoptosis induction in cancer cells by Allium vegetable-derived organosulfur compounds: a review. Mutat Res 555:121–131
Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7:1070–1083
Khar A, Jakse J, Havey MJ (2008) Segregations for onion bulb colors reveal that red is controlled by at least three loci. J Am Soc Hortic Sci 133:42–47
Khosa JS, McCallum J, Dhatt AS, Macknight RC (2015) Enhancing onion breeding using molecular tools. Plant Breed 135:9–20
Kim S, Binzel M, Yoo K, Park S, Pike LM (2004a) Inactivation of DFR (dihydroflavonol 4-reductase) gene transcription results in blockage of anthocyanin production in yellow onions (Allium cepa). Mol Breed 14:253–263
Kim S, Binzel ML, Yoo K, Park S, Pike LM (2004b) Pink (P), a new locus responsible for a pink trait in onions (Allium cepa) resulting from natural mutations of anthocyanidin synthase. Mol Genet Genomics 272:18–27
Kim S, Jones R, Yoo K, Pike LM (2005a) The L locus, one of complementary genes required for anthocyanin production in onions (Allium cepa), encodes anthocyanidin synthase. Theor Appl Genet 111:120–127
Kim S, Yoo K, Pike LM (2005b) The basic color factor, the C locus, encodes a regulatory gene controlling transcription of chalcone synthase genes in onions (Allium cepa). Euphytica 142:273–282
Kim S, Yoo K, Pike LM (2005c) Development of a PCR-based marker utilizing a deletion mutation in the DFR (dihydroflavonol 4-reductase) gene responsible for the lack of anthocyanin production in yellow onions (Allium cepa). Theor Appl Genet 110:588–595
Kim S, Bang H, Yoo K, Pike LM (2006) Identification of the fourth allele of the ANS (anthocyanidin synthase) gene and its effect on red color intensity in onions (Allium cepa). Euphytica 149:45–51
Kim S, Baek D, Cho DY, Yoon M (2009) Identification of two novel inactive DFR-A alleles responsible for failure to produce anthocyanin and development of a simple PCR-based molecular marker for bulb color selection in onion (Allium cepa L.). Theor Appl Genet 118:1391–1399
Kim S, Park JY, Yang T (2015) Characterization of three active transposable elements recently inserted in three independent DFR-A alleles and one high-copy DNA transposon isolated from the Pink allele of the ANS gene in onion (Allium cepa L.). Mol Genet Genomics 290:1027–1037
Lotito SB, Frei B (2006) Comsumption of flavonoids-rich foods and increased plasma antioxidant capacity in humans: cause, consequence, or epiphenomenon? Free Radic Biol Med 41:1727–1746
Neu-Yilik G, Gehring NH, Hentze MW, Kulozik AE (2004) Nonsense-mediated mRNA decay: from vacuum cleaner to Swiss army knife. Genome Biol 5:218
Nishiumi S, Miyamoto S, Kawabata K, Ohnishi K, Mukai R, Murakami A, Ashida H, Terao J (2011) Dietary flavonoids as cancer-preventive and therapeutic biofactors. Front Biosci 3:1332–1362
Park J, Cho DY, Moon JS, Yoon M, Kim S (2013) Development of functional markers for detection of inactive DFR-A alleles responsible for failure of anthocyanin production in onions (Allium cepa L.). Korean J Hortic Sci Technol 31:72–79
Petroni K, Tonelli C (2011) Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci 181:219–229
Quattrocchio F, Wing JF, Leppen HTC, Mol JN, Koes RE (1993) Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell 5:1497–1512
Ramsay NA, Glover BJ (2005) MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 10:63–70
Reiman GH (1931) Genetic factors for pigmentation in the onion and their relation to disease resistance. J Agric Res 42:251–278
Rhodes MJC, Price KR (1996) Analytical problems in the study of flavonoid compounds in onions. Food Chem 57:113–117
Ricroch A, Yockteng R, Brown SC, Nadot S (2005) Evolution of genome size across some cultivated Allium species. Genome 48:511–520
Russo M, Spagnuolo C, Tedesco I, Bilotto S, Russo GL (2012) The flavonoid quercetin in disease prevention and therapy: facts and fancies. Biochem Pharmacol 83:6–15
Shaul O (2015) Unique aspects of plant nonsense-mediated mRNA decay. Trends Plant Sci 20:767–779
Shirley BW (1996) Flavonoid biosynthesis: ‘new’ functions for an ‘old’ pathway. Trends Plant Sci 1:377–382
Slimestad R, Fossen T, Vågen IM (2007) Onions: a source of unique dietary flavonoids. J Agric Food Chem 55:10067–10080
Song S, Kim C, Moon JS, Kim S (2014) At least nine independent natural mutations of the DFR-A gene are responsible for appearance of yellow onions (Allium cepa L.) from red progenitors. Mol Breed 33:173–186
Spelt C, Quattrocchio F, Mol JN, Koes RE (2000) anthocyanin1 of Petunia encodes a basic helix–loop–helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 12:1619–1631
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Veitch NC, Grayer RJ (2011) Flavonoids and their glycosides, including anthocyanins. Nat Prod Rep 28:1626–1695
Vogt T (2010) Phenylpropanoid biosynthesis. Mol Plant 3:2–20
Wilmouth RC, Turnbull JJ, Welford RWD, Clifton IJ, Prescott AG, Schofield CJ (2002) Structure and mechanism of anthocyanidin synthase from Arabidopsis thaliana. Structure 10:93–103
Yamazaki M, Makita Y, Springob K, Saito K (2003) Regulatory mechanisms for anthocyanin biosynthesis in chemotypes of Perilla frutescens var. crispa. Biochem Eng J 14:191–197
Acknowledgments
This research was supported by the Agriculture Research Center Program, Golden Seed Project (Center for Horticultural Seed Development, No. 213008-04-4-SB910) and a Grant from the Next-Generation BioGreen 21 Program (Plant Molecular Breeding Center No. PJ011034). The authors thank Ji-wha Hur, Jeong-Ahn Yoo, and Su-jung Kim for their dedicated technical assistance.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10681_2016_1774_MOESM5_ESM.tif
Supplementary material 5 (TIFF 235) kb Supplementary Fig. 1. Phylogenetic tree constructed using amino acid sequences of 53 ANS enzymes isolated from different species. The GenBank accession numbers are shown in parenthesis. The numbers at the nodes are the bootstrap probability (%) with 1000 replicates. The scale bars indicate nucleotide substitutions per site
10681_2016_1774_MOESM6_ESM.tif
Supplementary material 6 (TIFF 508) kb Supplementary Fig. 2. Onion bulbs of the four red F1 hybrids between a yellow cultivar, ‘Santero’ and four yellow breeding lines. 1145 ‘Santero’ × OB814, 1146 ‘Santero’ × OB251, 1147 ‘Santero’ × OB252, 1148 ‘Santero’ × OB870
10681_2016_1774_MOESM7_ESM.tif
Supplementary material 7 (TIFF 113) kb Supplementary Fig. 3. Alignment of amino acid sequences of ANS enzymes isolated from Arabidopsis thaliana (NP194019) and three onion variants. ‘G229R’, ‘S188L’, and ‘L’ represent ANS G229R, ANS S188L, ANS L alleles, respectively. The positions of G229 and S188 are indicated with vertical arrows. Binding sites of iron and 2-oxoglutarate are indicated with filled and empty circles, respectively, and binding sites of dihydroquercetin and MES/ascorbate are indicated with filled and empty inverted triangles, respectively
10681_2016_1774_MOESM8_ESM.tif
Supplementary material 8 (TIFF 445) kb Supplementary Fig. 4. Alignment of amino acid sequences of 52 ANS enzymes isolated from other plant species and two onion ANS enzymes encoded by ANS P-1 and ANS h1-4 alleles. Only partial alignments around the positions containing amino acid changes among two onion ANS enzymes are presented. The dotted lines indicate the omitted regions. The two positions containing amino acid changes are enclosed with empty rectangular boxes
10681_2016_1774_MOESM9_ESM.tif
Supplementary material 9 (TIFF 350) kb Supplementary Fig. 5. Alignment of nucleotide sequences of the ANS h1, ANS h1-4, and ANS P alleles showing origination of the ANS h1-4 allele by intragenic recombination between ANS h1 and ANS P alleles. Two identical nucleotides at the polymorphic sequences among three ANS alleles are enclosed with empty rectangular boxes. The intron sequences are underlined
Rights and permissions
About this article
Cite this article
Kim, EY., Kim, CW. & Kim, S. Identification of two novel mutant ANS alleles responsible for inactivation of anthocyanidin synthase and failure of anthocyanin production in onion (Allium cepa L.). Euphytica 212, 427–437 (2016). https://doi.org/10.1007/s10681-016-1774-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-016-1774-3