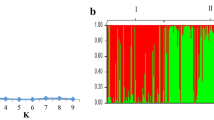
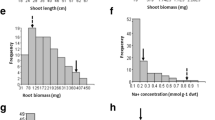
Abstract
Salinity is globally a major constraint for crop production. Breeding for salinity tolerance is an effective approach to improve crop production and productivity under saline conditions provided it is based on a good understanding of the genetic control of salinity tolerance. This study deals with mapping QTLs for salinity tolerance in durum wheat (Triticum durum) by association analysis using SSR markers. A total of 119 varieties were treated in 100 mM of NaCl solution and the salinity tolerance indices (STI) for several traits were calculated as parameters to assess salinity tolerance. Among the traits assessed, the increased proportion of dead leaves (%DL) was the most suitable parameter for assessment of salinity tolerance in durum varieties at early vegetative stages because of a broader range of variation among varieties and narrower range of variation within varieties compared to other traits. The QTL associated with salinity tolerance using %DL as a parameter was detected on chromosome 4B. An additional 11 QTLs associated with seven parameters using STI of other traits were detected on chromosomes 3A, 5A, 5B, 6A and 7A.


Similar content being viewed by others
Abbreviations
- QTL:
-
Quantitative trait locus
- SSR:
-
Simple sequence repeats
- STI:
-
Salt tolerance index
- EC:
-
Electrical conductivity
- LD:
-
Linkage disequilibrium
- GLM:
-
General linear model
- MLM:
-
Mixed linear model
- CHL:
-
Chlorophyll content
- NT:
-
Number of tillers per plant
- NL:
-
Number of leaves per tiller
- LL:
-
Leaf length
- TFW:
-
Total fresh weight of shoot and root
- SL:
-
Shoot length
- RL:
-
Root length
- SDW:
-
Shoot dry weight
- RDW:
-
Root dry weight
- %DL:
-
Increased percentage of dead leaves
- NFS:
-
Number of fertile spikes
- PH:
-
Plant height
- BIO:
-
Biomass production
- NS:
-
Number of seeds per spike
References
Anderson JA, Churchill JE, Autrique SD, Tanksley S, Sorrells ME (1993) Optimizing parental selection for genetic linkage maps. Genome 36:181–188
Ashraf M, O’Leary JW (1996) Responses of newly developed salt-tolerant genotype of spring wheat to salt stress: yield components and ion distribution. J Agron Crop Sci 176:91–101
Ayers RS, Westcot DW (1985) Water quality for agriculture, FAO Irrigation and Drainage Paper 29. Food and Agriculture Organization (FAO) of the United Nations, Rome
Azhar FM, McNeilly T (1989) The response of four sorghum varieties/cultivars to salinity during plant development. J Agron Crop Sci 163:33–43
Belaid A (2000) Durum wheat in WANA (West Asia and North Africa): production, trade, and gains from technological change. In: Royo C, Nachit MM, Di Fonzo N, Araus JL (eds) Durum wheat improvement in the Mediterranean region: new challenges, vol 40., Options MediterraneennesCIHEAM-IAMZ, Zaragoza, pp 35–39
Breseghello F, Sorrells ME (2006) Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 172:1165–1177
Byrt CS, Platten JD, Spielmeyer W, James RA, Lagudah ES, Dennis ES, Tester M, Munns R (2007) HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiol 143:1918–1928
Crossa J, Burgueno J, Dreisigacker S, Vargas M, Herrera Foessel SA, Lillemo M, Singh RP, Trethowan R, Warburton M, Franco J, Reynolds M, Crouch JH, Ortiz R (2007) Association analysis of historical bread wheat germplasm using additive genetic covariance of relatives and population structure. Genetics 177:1889–1913
Davenport R, James RA, Zakrisson-Plogander A, Tester M, Munns R (2005) Control of sodium transport in durum wheat. Plant Physiol 137:807–818
Diaz De Leon JL, Escoppinichi R, Geraldo N, Castellanos T, Mujeeb_Kazi A, Roder MS (2011) Quantitative parameter loci associated with salinity tolerance in field grown bread wheat. Euphytica 181:371–383
Dubcovsky J, Santa María G, Epstein E, Luo MC, Dvorak J (1996) Mapping of the K+/Na+ discrimination locus Kna1 in wheat. Theor Appl Genet 92:448–454
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Flint-Garcia SA, Thuillet AC, Yu JM, Pressoir G, Romero SM, Mitchell SE, Doebley J, Kresovich S, Goodman MM, Buckler ES (2005) Maize association population: a high resolution platform for quantitative parameter locus dissection. Plant J 44:1054–1064
Flowers TJ, Garcia A, Koyama M, Yeo AR (1997) Breeding for salt tolerance in crop plants—the role of molecular biology. Acta Physiol Plant 19:427–433
Fu D, Szucs P, Yan L, Helguera M, Skinner JS, von Zitzewitz J, Hayes PM, Dubcovsky J (2005) Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol Genet Genomics 273:54–65
Garcia-Suarez JV, Diaz De Leon JL, Roder MS (2010) Identification of QTLs and associated molecular markers related to starch degradation in wheat seedlings (Triticum aestivum L.) under saline stress. Cereal Res Commun 38:163–174
Gaut BS, Long AD (2003) The lowdown on linkage disequilibrium. Plant Cell 15:1502–1506
Genc Y, Oldach K, Verbyla A, Lott G, Hassan M, Tester M, Wallwork H, McDonald G (2010) Sodium exclusion QTL associated with improved seedling growth in bread wheat under salinity stress. Theor Appl Genet 121:877–894
Goudarzi M, Pakniyat H (2008) Evaluation of wheat cultivars under salinity stress based on some agronomic and physiological traits. J Agric Soc Sci 1:35–38
Gupta PK (2002) Molecular markers and QTL analysis in crop plants. Curr Sci 83:113–114
Gupta PK, Rustgi S, Kulwal PL (2005) Linkage disequilibrium and association studies in higher plants: present status and future prospects. Plant Mol Biol 57:461–485
Guyomarc’h H, Sourdille P, Charmet G, Edwards J, Bernard M (2002) Characterisation of polymorphic microsatellite markers from Aegilops tauschii and transferability to the D-genome of bread wheat. Theor Appl Genet 104:1164–1172
Hagemann M, Erdmann N (1997) Environmental stresses. In: Rai AK (ed) Cyanobacterial nitrogen metabolism and environmental biotechnology. Springer, Heidelberg, pp 156–221
Hayashi H, Murata N (1998) Genetically engineered enhancement of salinity tolerance in higher plants. In: Satoh K, Murata N (eds) Stress response of photosynthetic organisms: molecular mechanisms and molecular regulation. Elsevier, Amsterdam, pp 133–148
Hernandez JA, Almansa MS (2002) Short-term effects of salt stress on antioxidant systems and leaf water relations of pea leaves. Physiol Plant 115:251–257
Hirschhorn JN, Daly MJ (2005) Genome-wide association studies for common diseases and complex traits. Nat Rev Genet 6:95–108
James RA, Davenport RJ, Munns R (2006) Physiological characterisation of two genes for Na+ exclusion in durum wheat: Nax1 and Nax2. Plant Physiol 142:1537–1547
Kato K, Miura H, Sawada S (2000) Mapping QTL controlling grain yield and its components on chromosome 5A of wheat. Theor Appl Genet 101:1114–1121
Koebner RMD, Martin PK, Orford SM, Miller TE (1996) Responses to salt stress controlled by the homeologous group 5 chromosomes of hexaploid wheat. Plant Breed 115:81–84
Lindsay MP, Lagudah ES, Hare RA, Munns R (2004) A locus for sodium exclusion (Nax1), a trait for salt tolerance, mapped in durum wheat. Funct Plant Biol 31:1105–1114
Liu K, Muse SV (2005) Power maker: an integrated analysis environment for genetic maker analysis. Bioinformatics 21:2128–2129
Ma L, Zhou E, Huo N, Zhou R, Wang G, Jia J (2007) Genetic analysis of salinity tolerance in a recombinant inbred population of wheat (Triticum aestivum L.). Euphytica 153:109–117
Maas EV, Poss JA, Hoffman GJ (1986) Salinity tolerance of sorghum at three growth stages. Irrig Sci 7:1–11
Morancho J (1995) World durum wheat trade. In: Di Fonzo N, Kaan F, Nachit M (eds) Durum wheat quality in the Mediterranean region, vol 22., Options MediterraneennesCIHEAM-IAMZ, Zaragoza, pp 213–219
Munns R (1993) Physiological processes limiting plant growth in saline soils: some dogmas and hypotheses. Plant Cell Environ 16:15–24
Munns R, James R (2003) Screening methods for salinity tolerance: a case study with tetraploid wheat. Plant Soil 253:201–218
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
Munns R, James RA, Xu B, Athman A, Conn SJ, Jordans C, Byrt CS, Hare RA, Tyerman SD, Tester M, Plett D, Gilliham M (2012) Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat Biotechnol 30:360–364
Noori SAS, McNeilly T (2000) Assessment of variability in salt tolerance based on seedling growth in Triticum durum Desf. Genet Resour Crop Evol 47:285–291
Nordborg M, Borevitz JO, Bergelson JO, Berry CC, Chory J, Hagenblad J, Kreitman M, Maloof JN, Noyes T, Stahl E, Oefner PJ, Stahl E, Weigel D (2002) The extent of linkage disequilibrium in the highly selfing species Arabidopsis thaliana. Nat Genet 30:190–193
Parida A, Das A (2005) Salinity tolerance and salinity effects on plants: a review. Ecotoxicol Environ Saf 60:324–349
Pearson GA, Bernstein L (1959) Salinity effects at several growth stages of rice. Agron J 51:654–657
Pestsova E, Korzun V, Goncharov NP, Hammer K, Ganal MW, Rodder MS (2000) Microsatellite analysis of Aegilops tauschii germplasm. Theor Appl Genet 101:100–106
Poustini K, Siosemardeh A (2004) Ion distribution in wheat cultivars in response to salinity stress. Field Crop Res 85:125–133
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rashid A, Qureshi RH, Hollington PA, Wyn Jones RG (1999) Comparative response of wheat (Triticum aestivum L.) cultivars to salinity at the seedling stage. Agron Crop Sci 182:199–207
Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, Kresovich S, Goodman MM, Buckler ES IV (2001) Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc Natl Acad Sci USA 98:11479–11484
Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal MW (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Schachtman DP, Lagudah ES, Munns R (1992) The expression of salt tolerance from Triticum tauschii in hexaploid wheat. Theor Appl Genet 84:714–719
Singh SB, Singh BB, Singh M (1994) Effect of kinetin on chlorophyll, nitrogen and proline in mungbean under saline conditions. Indian J Plant Physiol 37:37–39
Skøt L, Humphreys J, Humphreys MO, Thorogood D, Gallagher J, Sanderson R, Armstead IP, Thomas ID (2007) Association of candidate genes with flowering time and water-soluble carbohydrate content in Lolium perenne (L.). Genetics 177:535–547
Somers DJ, Isaac P, Edwards K (2004) A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109:1105–1114
Somers DJ, Banks T, DePauw R, Fox S, Clarke J, Poznial C, McCartney C (2007) Genome-wide linkage disequilibrium analysis in bread wheat and durum wheat. Genome 50:557–567
Song QJ, Fickus EW, Cregan PB (2002) Characterization of trinucleotide SSR motifs in wheat. Theor Appl Genet 104:286–293
Song QJ, Shi JR, Singh S, Fickus EW, Costa JM, Lewis J, Gill BS, Ward R, Cregan PB (2005) Development and mapping of microsatellite (SSR) markers in wheat. Theor Appl Genet 110:550–560
Sourdille P, Cadalen T, Guyomarc’h H, Snape JW, Perretant MR, Charmet G, Boeuf C, Bernard S, Bernard M (2003) An update of the Courtot×Chinese Spring intervarietal molecular marker linkage map for the QTL detection of agronomic traits in wheat. Theor Appl Genet 106:530–538
Stich B, Maurer HP, Melchinger AE, Frisch M, Heckenberger M, Rouppe van der Voort J, Peleman J, Sørensen AP, Reif JC (2006) Comparison of linkage disequilibrium in elite European maize inbred lines using AFLP and SSR markers. Mol Breed 17:217–226
Stich B, Mohring J, Piepho HP, Heckenberger M, Buckler ES, Melchinger AE (2008) Comparison of mixed-model approaches for association mapping. Genetics 178:1745–1754
Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot 91:503–527
Tommasini L, Schnurbusch T, Fossati D, Mascher F, Keller B (2007) Association mapping of Stagonospora nodorum blotch resistance in modern European winter wheat varieties. Theor Appl Genet 115:697–708
Tuteja N (2007) Mechanisms of high salinity tolerance in plants. Methods Enzymol 428:419–438
USDA-ARS (2008) Research databases. Bibliography on salinity tolerance. George E Brown Jr. Salinity Lab, US Dept Agric, Agric, Res, Serv, Riverside, CA. http://www.ars.usda.gov/Services/docs.htmdocid=8908
Weir BS (1996) Genetic data analysis II. Methods for discrete population genetic data. Sinauer Associates, Sunderland
Xu Y, Li S, Li L, Zhang X, Xu H, An D (2013) Mapping QTLs for salinity tolerance with additive, epistatic and QTL× treatment interaction effects at seedling stage in wheat. Plant Breed 132:276–280
Yu H, Deng Z, Xiang C, Tian J (2013) Analysis of diversity and linkage disequilibrium mapping of agronomic traits on B-genome of wheat. Genomics 2:20–30
Acknowledgments
This research was carried out by GRANDE project supported by JST/JICA, SATREPS (Science and Technology Research Partnership for Sustainable Development), Japan.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Turki, N., Shehzad, T., Harrabi, M. et al. Detection of QTLs associated with salinity tolerance in durum wheat based on association analysis. Euphytica 201, 29–41 (2015). https://doi.org/10.1007/s10681-014-1164-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-014-1164-7