Summary
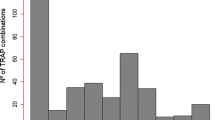
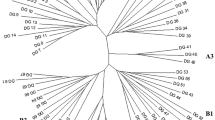
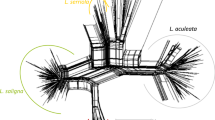
To demonstrate the applicability of the target region amplification polymorphism (TRAP) marker technique to lettuce genotyping, we fingerprinted 53 lettuce (Lactuca sativa L.) cultivars and six wild accessions (three from each of the two wild species, L. saligna L. and L. serriola L.). Seven hundred and sixty-nine fragments from 50 to 900 bp in length were amplified in 10 PCR reactions using 10 fixed primers in combination with four fluorescent labeled arbitrary primers. Three hundred and eighty-eight of these fragments were polymorphic among the 59 Lactuca entries and 107 fragments were polymorphic among the 53 lettuce cultivars and the six wild accessions; 251 fragments were present only in the wild species. These markers not only discriminated all cultivars, but also revealed the evolutionary relationship among the three species: L. sativa, the cultivated species, is more closely related to L. serriola than to L. saligna. Cluster analysis grouped the cultivars by horticultural types with a few exceptions. These results are consistent with previous findings using RFLP, AFLP, and SAMPL markers. The TRAP markers revealed significant differences in genetic variability among horticultural types, measured by the average genetic similarity among the cultivars of the same type. Within the sample set, the leaf type and butterhead types possessed relatively high genetic variability, the iceberg types had moderate variability and the romaine types had the lowest variability. The genetic behavior of TRAP markers was assessed with a mapping population of 45 recombinant inbred lines (RILs) derived from an interspecific cross between L. serriola and L. sativa. Almost all the markers segregated in the expected 1:1 Mendelian ratio and are being incorporated into the existing lettuce linkage maps. Our results indicate that the TRAP markers can provide a powerful technique for fingerprinting lettuce cultivars.
Similar content being viewed by others
References
Cho, H.S. & H.S. Pai, 2000. Cloning and characterization of ntTMK1 gene encoding a TMK1-homologous receptor-like kinase in tobacco. Mol Cells 10: 317–24.
Deshpande, A.D., W. Ramakrishna, G.P. Mulay, V.S. Gupta & P.K. Ranjekar, 1998. Evolutionary and polymorphic organization of the knotted1 homeobox in cereals. Theor Appl Genet 97: 135–140.
Dice, L.R., 1945. Measures of the amount of ecologic association between species. Ecology 26: 297–302.
Gehring, W.J., 1987. Homeoboxes in the study of development. Science 236: 1245–1252.
Glase, L., G. Lucier & G. Thompson, 2001. Lettuce: in and out of the bag. Agricultural Outlook/April 2001. Website: http://www.ers.usda.gov/publications/AgOutlook/April2001/AO280d.pdf.
Hagen, L., B. Khadari, P. Lambert & J.M. Audergon, 2002. Genetic diversity in apricot revealed by AFLP markers: Species and cultivar comparisons. Theor Appl Genet 105: 298–305.
Hill, M., H. Witsenboer, M. Zabeau, P. Vos, R. Kesseli & R.W. Michelmore, 1996. AFLP fingerprinting as a tool for studying genetic relationships in Lactuca spp. Theor Appl Genet 93: 1202–1210.
Hoffman, D., A. Hang, S. Larson & B. Jones, 2003. Conversion of an RAPD marker to an STS marker for barley variety identification. Plant Mol Biol Reporter 21: 81–91.
Hu, J. & B.A. Vick, 2003. TRAP (target region amplification polymorphism), a novel marker technique for plant genotyping. Plant Mol Biol Reporter 21: 289–294.
Hu, J., J. Chen, A. Berville & BA. Vick, 2005. Defining sunflower linkage group ends with the conserved telomere sequence-derived TRAP markers. Plant Genome XIII. January 15–19, San Diego, CA.
Jeuken, M., R. van Wijk, J. Peleman & P. Lindhout, 2001. An integrated interspecific AFLP map of lettuce (Lactuca) based on two L. sativa × L. saligna F2 populations. Theor Appl Genet 103: 638–647.
Kesseli, R.V. & R.W. Michelmore, 1986. Genetic variation and phylogenies detected from isozyme markers in species of Lactuca. J. Hered. 77: 324–331.
Kesseli, R.V., I. Paran & R.W. Michelmore, 1994. Analysis of a detailed genetic linkage map of Lactuca sativa (lettuce) constructed from RFLP and RAPD markers. Genetics 136: 1435–1446.
Kesseli, R.V., O.E. Ochoa & R.W. Michelmore, 1991. Variation at RFLP loci in Lactuca spp. and origin of cultivated lettuce. Genome 34: 430–436.
Landry, P.A. & F.J. Lapointe, 1996. RAPD problems in phylogenetics. Zoologica Scripta 25: 283–290.
Li, G. & C.F. Quiros, 2001. Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theor Appl Genet 103: 455–461.
Liu, Z., J.A. Anderson, J. Hu, T.L. Friesen, J.B. Rasmussen & J.D. Faris, 2005. A wheat intervarietal linkage map based on microsatellite and target region amplified polymorphism markers and its utility for detecting quantitative trait loci. Theor Appl Genet (in press).
McGinnis, W., R.L. Garber, J. Wirz, A. Kuroiwa & W.J. Gehring, 1984. A homologous protein-coding sequence in Drosophila homeotic genes and its conservation in other metazoans. Cell 37: 403–408.
Pujar, S., S.A. Tamhankar, V.S. Rao, V.S. Gupta, S. Naik & P.K. Ranjekar, 1999. Arbitrarily primed-PCR based diversity assessment reflects hierarchical groupings of Indian tetraploid wheat genotypes. Theor Appl Genet 99: 868–876.
Rozen, S. & H.J. Skaletsky, 2000. Primer3 on the WWW for general users and for biologist programmers. In: S. Krawetz & S. Misener (Eds.) Bioinformatics Methods and Protocols: Methods in Molecular Biology, pp. 365–386. Humana Press, Totowa, NJ.
Sakuoka, R., R.T. Hamasaki, R. Shimabuku & A. Arakaki, 2000. Lettuce for the home garden. CTAHR's Home Garden Vegetable Series. Website: http://www2.ctahr.hawaii.edu/oc/freepubs/pdf/HGV-2.pdf.
Sambrook, J., E.F. Fritsch & T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual, 2nd edn., Cold Spring Harbor Laboratory, New York.
Scott, M.P., J.W. Tamkun & G.W. Hartzell, 1989. The structure and function of the homeodomain. BBA Reviews on Cancer 989: 25–48.
Sneath, P.H.A. & R.R. Sokal, 1973. Numerical Taxonomy: The Principal and Practice of Numerical Classification. W. H. Freeman and Company, San Francisco.
UC Davis Compositae Database: Website: http://compositdb.ucdavis.edu/.
van Stallen, N., V. Noten, M. Demeulemeester & M. de Proft, 2000. Identification of commercial chicory cultivars for hydroponic forcing and their phenetic relationships revealed by random amplified polymorphic DNAs and amplified fragment length polymorphisms. Plant Breed 119: 265–270.
Vollbrecht, E., B. Veit, N. Sinha & S. Hake, 1991. The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 350: 241–243.
Witsenboer, H., J. Vogel & R.W. Michelmore, 1998. Identification, genetic localization and allelic diversity of selectively amplified microsatellite polymorphic loci (SAMPL) in lettuce and wild relatives (Lactuca spp.). Genome 40: 923–936.
Author information
Authors and Affiliations
Corresponding author
Additional information
The U.S. Government's right to retain a non-exclusive, royalty-free license in and to any copyright is acknowledged.
Rights and permissions
About this article
Cite this article
Hu, J., Ochoa, O.E., Truco, M.J. et al. Application of the TRAP technique to lettuce (Lactuca sativa L.) genotyping. Euphytica 144, 225–235 (2005). https://doi.org/10.1007/s10681-005-6431-1
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s10681-005-6431-1