Abstract
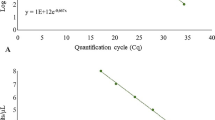
Pythium inflatum is the causal agent of Pythium maize stalk rot, which is one of the most devastating diseases of maize (Zea mays L.). P. inflatum is currently a major concern in global maize production. To the best of our knowledge, no effective resistance to P. inflatum is known in maize, and no effective measures have been reported for the control of this pathogen once maize plants have been infected. Early and accurate detection of P. inflatum is essential to guide maize planting and to protect maize production. A real-time fluorescence loop-mediated isothermal amplification (RealAmp) assay was developed for the rapid quantitative detection of P. inflatum in soil. The detection limit of the RealAmp assay was approximately 0.1 pg/μl plasmid DNA when mixed with extracted soil DNA or 103 spores/g of artificially infested soil. No cross-reactions with other related pathogens were observed. Results of the RealAmp assay for quantifying the genomic DNA of P. inflatum were confirmed by testing with artificially and naturally infested samples. The quantification of the soil-borne pathogen DNA of P. inflatum in naturally infested samples was not significantly different compared with classic real-time PCR (P < 0.05). Additionally, the RealAmp assay could be detected via an improved closed-tube visual detection system by adding SYBR Green I fluorescent dye to the inside of the lid prior to amplification. Consequently, the inhibitory effects of the stain on DNA amplification were avoided. Therefore, the assay could be used more conveniently in the field as a simple, rapid, and effective technique and has the potential to become an alternative tool for detecting and monitoring P. inflatum in the field.




Similar content being viewed by others
References
Abdelzaher, H. M. A., Ichitani, T., & Elnaghy, M. A. (1994). Effect of temperature, hydrogen ion concentration and osmotic potential on oospore germination of five Pythium spp. isolated from pond water. Mycoscience, 35, 315–318.
Agrios, G. N. (2005). Plant pathology (5th ed.). Burlington: Elsevier academic press.
Asano, T., Senda, M., Suga, H., & Kageyama, K. (2010). Development of multiplex PCR to detect five Pythium species related turfgrass disease. Journal of Phytopathology, 158, 609–615.
Ayers, W. A., & Lumsden, R. D. (1975). Factors affecting production and germination of oospores of three Pythium species. Phytopathology, 65, 1094–1100.
Bahramisharif, A., Lamprecht, S. C., Spies, C. F. J., Botha, W. J., Calitz, F. J., & McLeod, A. (2014). Pythium spp. associated with rooibos seedlings, and their pathogenicity toward rooibos, lupin, and oat. Plant Disease, 98, 223–242.
Bekele, B., Hodgetts, J., Tomlinson, J., Boonham, N., Nikolic, P., Swarbrick, P., & Dickinson, M. (2011). Use of a real-time LAMP isothermal assay for detecting 16SrII and XII phytoplasmas in fruit and weeds of the Ethiopian Rift Valley. Plant Pathology, 60, 345–355.
Broders, K. D., Lipps, P. E., Paul, P. A., & Dorrance, A. E. (2007a). Charcterization of Pythium spp. associated with corn and soybean seed and seedling disease in Ohio. Plant Disease, 91, 727–731.
Broders, K. D., Lipps, P. E., Paul, P. A., & Dorrance, A. E. (2007b). Evaluation of Fusarium graminearum associated with corn and soybean seed and seedling disease in Ohio. Plant Disease, 91, 1155–1160.
Chen, W., & Hoy, J. W. (1993). Molecular and morphological comparison of Pythium arrhenomanes and P. graminicola. Mycological Research, 97, 1371–1378.
Dick, M. W. (1990). Keys to Pythium (pp. 1–64). UK: Department of Botany, School of Plant Sciences, University of Reading.
Doyle, J. J., & Doyle, J. L. (1990). Isolation of plant DNA from fresh tissue. Focus, 12, 13–15.
Fukuta, S., Takahashi, R., Kuroyanagi, S., Miyake, N., Nagai, H., Suzuki, H., Hashizume, F., Tsuji, T., Taguchi, H., Watanabe, H., & Kageyama, K. (2013). Detection of Pythium aphanidermatum in tomato using loop-mediated isothermal amplification (LAMP) with species-specific primers. European Journal of Plant Pathology, 136, 689–701.
Fukuta, S., Takahashi, R., Kuroyanagi, S., Ishiguro, Y., Miyake, N., Nagai, H., Suzuki, H., Tsuji, T., Hashizume, F., Watanabe, H., & Kageyama, K. (2014). Development of loop-mediated isothermal amplification assay for the detection of Pythium myriotylum. Letters in Applied Microbiology, 59, 49–57.
Godfrey, S. A., Monds, R. D., Lash, D. T., & Marshall, J. W. (2003). Identification of Pythium oligandrum using species-specific ITS rDNA PCR oligonucleotides. Mycological Research, 107, 790–796.
Góomez-Alpízar, L., Saalau, E., Picado, I., Tambong, J. T., & Saborío, F. (2011). A PCR-RFLP assay for identification and detection of Pythium myriotylum, causal agent of the cocoyam root rot disease. Letters in Applied Microbiology, 52, 185–192.
Goto, M., Honda, E., Ogura, A., Notomo, A., & Hanaki, K. I. (2009). Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques, 46, 167–172.
He, J., Guo, Q., Wang, X., Song, L., Zhang, W., & Wu, X. (2011). Study on genetic diversity of Fusarium graminearum populations causing maize stalk rot by ISSR analysis. Journal of Maize Science, 19, 129–134.
Ishiguro, Y., Asano, T., Otsubo, K., Suga, H., & Kageyama, K. (2013). Simultaneous detection by multiplex PCR of the high-temperature-growing Pythium species: P. aphanidermatum, P. helicoides and P. myriotylum. Journal of General Plant Pathology, 79, 350–358.
Iwamoto, T., Sonobe, T., & Hayashi, K. (2003). Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. Journal of Clinical Microbiology, 41, 2616–2622.
Kageyama, K. (2014). Molecular taxonomy and its application to ecological studies of Pythium species. Journal of General Plant Pathology. doi:10.1007/s10327-014-0526-2.
Kageyama, K., Ohyama, A., & Hyakumachi, M. (1997). Detection of Pythium ultimum using polymerase chain reaction with species specific primers. Plant Disease, 81, 1155–1160.
Kageyama, K., Nakashima, A., Kajihara, Y., Suga, H., & Nelson, E. B. (2005). Phylogenetic and morphological analyses of Pythium graminicola and related species. Journal of General Plant Pathology, 71, 174–182.
Kernaghan, G., Reeleder, R. D., & Hoke, S. M. T. (2008). Quantification of Pythium populations in ginseng soils by culture dependent and real-time PCR methods. Applied Soil Ecology, 40, 447–455.
Kiatpathomchai, W., Jaroenram, W., Arunrut, N., Jitrapakdee, S., & Flegel, T. W. (2008). Shrimp Taura syndrome virus detection by reverse transcription loop-mediated isothermal amplification combined with a lateral flow dipstick. Journal of Virological Methods, 153, 214–217.
Le Floch, G., Tambong, J., Vallance, J., Tirilly, Y., Levesque, A., & Rey, P. (2007). Rhizosphere persistence of three Pythium oligandrum strains in tomato soilless culture assessed by DNA microarray and real-time PCR. FEMS Microbiology Ecology, 61, 317–326.
Lévesque, C. A., & de Cock, A. W. (2004). Molecular phylogeny and taxonomy of the genus Pythium. Mycological Research, 108, 1363–1383.
Long, Y. Y., Wei, J. G., Huang, C. L., He, Y. Q., Yuan, G. Q., Shi, Y., & Xiong, Y. (2010). A new Pythium species isolated from vegetable fields and analysis by rDNA ITS sequence. Mycosytema, 295, 795–800.
Lucchi, N. W., Demas, A., Narayanan, J., Sumari, D., Kabanywanyi, A., Kachur, S. P., Barnwell, J. W., & Udhayakumar, V. (2010). Real-time fluorescence loop mediated isothermal amplification for the diagnosis of malaria. PLoS ONE. doi:10.1371/journal.pone.0013733.
McLeod, A., Botha, W. J., Meitz, J. C., Spies, C. F. J., Tewoldemedhin, Y. T., & Mostert, L. (2009). Morphological and phylogenetic analyses of Pythium species in South Africa. Mycological Research, 113, 933–951.
Mori, Y., Nagamine, K., Tomita, N., & Notomi, T. (2001). Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochemical and Biophysical Research Communications, 289, 150–154.
Mori, Y., Kitao, M., Tomita, N., & Notomi, T. (2004). Real-time turbidimetry of LAMP reaction for quantifying template DNA. Journal of Biochemical and Biophysical Methods, 59, 145–157.
Nagamine, K., Watanabe, K., Ohtsuka, K., Hase, T., & Notomi, T. (2001). Loop-mediated isothermal amplification reaction using a nondenatured template. Clinical Chemistry, 47, 1742–1743.
Nagamine, K., Hase, T., & Notomi, T. (2002). Accelerated reaction by loop-mediated isothermal amplification using loop primers. Molecular and Cellular Probes, 16, 223–229.
Nechwatal, J., & Lebecka, R. (2014). Genetic and phenotypic analyses of Pythium isolates from reed suggest the occurrence of a new species, P. phragmiticola, and its involvement in the generation of a natural hybrid. Mycoscience, 55, 134–143.
Njiru, Z. K., Yeboah-Manu, D., Stinear, T. P., & Fyfed, J. A. (2012). Rapid and sensitive detection of Mycobacterium ulcerans by use of a loop-mediated isothermal amplification test. Journal of Clinical Microbiology, 50, 1737–1741.
Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., & Hase, T. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Research. doi:10.1093/nar/28.12.e63.
Ruano, G., Fenton, W., & Kidd, K. K. (1989). Biphasic amplification of very dilute DNA samples via ‘booster’ PCR. Nucleic Acids Research, 17(13), 5407.
Schroeder, K. L., Martin, F. N., de Cock, A. W. A. M., Lévesque, C. A., Spies, C. F. J., Okubara, P. A., & Paulitz, T. C. (2013). Molecular detection and quantification of Pythium species: evolving taxonomy, new tools, and challenges. Plant Disease, 97, 4–20.
Tewoldemedhin, Y. T., Mazzola, M., Labuschagne, I., & McLeod, A. (2011). A multi-phasic approach reveals that apple replant disease is caused by multiple biological agents, with some agents acting synergistically. Soil Biology & Biochemistry, 43, 1917–1927.
Tomlinson, J. A., Dickinson, M. J., & Boonham, N. (2010). Rapid detection of Phytophthora ramorum and P. kernoviae by two-minute DNA extraction followed by isothermal amplification and amplicon detection by generic lateral flow device. Phytopathology, 100, 143–149.
Villa, N. O., Kageyama, K., Asano, T., & Suga, H. (2006). Phylogenetic relationships of Pythium and Phytophthora species based on ITS rDNA, cytochrome oxidase II and β- tubulin gene sequences. Mycologia, 98, 410–422.
Wang, P. H., & Chang, C. W. (2003). Detection of the low-germination-rate resting oospores of Pythium miriotylum from soil by PCR. Letters in Applied Microbiology, 36, 157–161.
Wang, X., Wu, Q., Liu, X., & Ma, G. (1994). Identification and pathogenicity of Pythium spp. isolated from maize. Acta Phytopathologica Sinica, 24, 343–346.
Wang, P. H., Boo, L. M., Lin, Y. S., & Yeh, Y. (2002). Specific detection of Pythium aphanidermatum from hydroponic nutrient solution by booster PCR with DNA primers developed from mitochondrial DNA. Phytoparasitica, 30, 473–485.
Wang, P. H., Chung, C. Y., Lin, Y. S., & Yeh, Y. (2003a). Use of polymerase chain reaction to detect the soft rot pathogen, Pythium myriotylum, in infected ginger rhizomes. Letters in Applied Microbiology, 36, 116–120.
Wang, P. H., Wang, Y. T., & White, J. G. (2003b). Species-specific PCR primers for Pythium developed from ribosomal ITS region. Letters in Applied Microbiology, 37, 127–132.
Wu, H., Sun, S., Fan, Z., Liu, C., Yang, T., & Zhu, J. (2007). Research condition and prevention counter measures of maize stalk rot. Journal of Maize Science, 15, 129–132.
Xu, S., Chen, J., Gao, Z., Ji, M., & Liu, H. (2006). Maize stalk and ear rot in China. Acta Phytopathologica Sinica, 36, 193–203.
Zhang, X., Zhang, H., Pu, J., Qi, Y., Yu, Q., Xie, Y., & Peng, J. (2014). Development of a real-time fluorescence loop-mediated isothermal amplification assay for rapid and quantitative detection of Fusarium oxysporum f. sp. cubense Tropical Race 4 in soil. PLoS ONE. doi:10.1371/annotation/8a1fd6a3-754f-42e2-a906-9d224167344e.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (#U1204320) and the HNAAS (Henan Academy of Agricultural Sciences) Foundation for Excellent Young Scholars (2013YQ03). We thank Professor Xiaoming Wang (Chinese Academy of Agricultural Sciences, Beijing, China) for providing the P. inflatum strain; Professor Jiguang Wei (Guangxi University, Nanning, China) for providing the P. graminicola, P. aphanidermatum, P. arrhenomanes, P. guangxiense, and P. periilum; and Dr. Junjie Hao (Henan Academy of Agricultural Sciences, Zhengzhou, China) for providing the F. graminearum.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(XLSX 21 kb)
ESM 2
(XLSX 15 kb)
Fig. S1
Specific identification of Pythium inflatum among Pythium spp. via the real-time fluorescence loop-mediated isothermal amplification (RealAmp) assay. a. Agarose gel electrophoresis analysis of RealAmp assay amplicons showing the specific test for P. inflatum. Lane 1 to lane 2, genomic DNAs of positive and negative controls, respectively; Lane 3 to lane 8, the DNAs of P. inflatum (HNAAS-19), P. inflatum (GuoLD001447), P. graminicola (CY-A156), P. acanthicum (CY-B424), P. arrhenomanes (CY-B386), and P. debaryanum, respectively. Lane M, D2000 DNA marker (Tiangen Biotech). The same samples were used for lane 1 to lane 8 of Fig. S1a to Fig. S1d. b. The RealAmp assay was validated by specific PCR amplification using the specific Pinf1/ITS2 primer set; Lane D, 50 bp DNA ladder (Tiangen Biotech). c. Visual inspection of the RealAmp amplification products. The original orange color of SYBR Green I turned green in the positive reaction mixture. d. The fluorescence vs. time graph was plotted automatically by the ESE-Quant Tube Scanner. The graph reports the fluorescence in mV on the y-axis and time in min on the x-axis. (GIF 35 kb)
Fig. S2
The sensitivity of Pythium inflatum real-time fluorescence loop-mediated isothermal amplification (RealAmp) test. a. Agarose gel electrophoresis analysis of RealAmp amplication products correspond to serial diluted plasmid DNA template. Lane 2 to lane 7 are 10-fold dilutions of the pPiITS plasmid DNA ranging from 10 ng/μl to 1.0 × 10-5 ng/μl. Lane 8, negative control. b. Visual detection of the RealAmp amplification products. The original orange color of SYBR Green I turned green in the positive reaction mixture. (GIF 19 kb)
Fig. S3
Screening of different sets of primers used for Pythium inflatum RealAmp reactions. The real-time fluorescence loop-mediated isothermal amplification assay (RealAmp) products were amplified from P. inflatum DNA (P1 and P2) and the DNA of other pathogens (N1 and N2). The fluorescence vs. time graph was plotted automatically by the ESE-Quant Tube Scanner. The graph reports the fluorescence in mV on the y-axis and time in min on the x-axis. (GIF 47 kb)
Rights and permissions
About this article
Cite this article
Cao, Y., Li, Y., Li, J. et al. Rapid and quantitative detection of Pythium inflatum by real-time fluorescence loop-mediated isothermal amplification assay. Eur J Plant Pathol 144, 83–95 (2016). https://doi.org/10.1007/s10658-015-0752-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-015-0752-2