Abstract
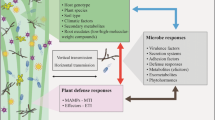
Plant viruses transmitted by the obligate root-infecting plasmodiophorid parasites Polymyxa graminis and Polymyxa betae cause devastating yield losses to cereal and sugar beet crops worldwide. Barley is a non-host for P. betae but is a host for P. graminis. Using the Barley1 GeneChip® microarray we have investigated the transcriptional re-programming of barley roots during the earliest non-host and host interactions with zoospores of these protist species. At high confidence levels we detected 20 and 13 genes with increased transcriptional activity in response to P. betae and P. graminis, respectively, compared to unchallenged barley roots. Functional classification of the induced genes showed that a majority of the genes from both responses were associated with a classic defence response. Validation by quantitative RT-PCR analysis indicated that all of the genes examined were induced to comparable levels in both non-host and host responses. Our results also demonstrated that the barley defence-associated genes, RAR1, ROR1 or ROR2 were not essential for limiting the establishment of P. betae infection in barley. These data suggest that in barley roots the Polymyxa species induce a similar basal defence response whether the interaction is with a non-host or host. Thus, the early response to protist plant parasites appears to be part of the general ‘frontline’ defence against invading microbes.



Similar content being viewed by others
Abbreviations
- ELISA:
-
enzyme-linked immunosorbent assay
- FDR:
-
false discovery rate
- PAMP:
-
pathogen-associated molecular pattern
- qRT-PCR:
-
quantitative reverse transcription polymerase chain reaction
References
Adams, M. J., & Swaby, A. G. (1988). Factors affecting the production and motility of zoospores of Polymyxa graminis and their transmission of Barley yellow mosaic virus (BaYMV). Annals of Applied Biology, 112, 69–78.
Barr, D. J. S. (1979). Morphology and host range of Polymyxa graminis, Polymyxa betae and Ligniera pilorum from Ontario and other areas. Canadian Journal of Plant Pathology, 1, 85–94.
Barr, K. J., Asher, M. J. C., & Lewis, B. G. (1995). Resistance to Polymyxa betae in wild Beta species. Plant Pathology, 44, 301–307.
Benjamini, Y., & Hochberg, Y. (1995). Controlling the false discovery rate—A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B, Statistical Methodology, 57, 289–300.
Burton, R. A., Shirley, N. J., King, B. J., Harvey, A. J., & Fincher, G. B. (2004). The CesA gene family of barley. Quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiology, 134, 224–236.
Caldo, R. A., Nettleton, D., Peng, J. Q., & Wise, R. P. (2006). Stage-specific suppression of basal defense discriminates barley plants containing fast- and delayed-acting Mla powdery mildew resistance alleles. Molecular Plant–Microbe Interactions, 19, 939–947.
Close, T. J. (2005). The barley microarray. A community vision and application to abiotic stress. Czech Journal of Genetics and Plant Breeding, 41, 144–152.
Close, T. J., Wanamaker, S. I., Caldo, R. A., Turner, S. M., Ashlock, D. A., Dickerson, J. A., et al. (2004). A new resource for cereal genomics: 22K barley GeneChip comes of age. Plant Physiology, 134, 960–968.
de Torres, M., Sanchez, P., Fernandez-Delmond, I., & Grant, M. (2003). Expression profiling of the host response to bacterial infection: the transition from basal to induced defence responses in RPM1-mediated resistance. Plant Journal, 33, 665–676.
Dimmer, E., Roden, L., Cai, D. G., Kingsnorth, C., & Mutasa-Göttgens, E. (2004). Transgenic analysis of sugar beet xyloglucan endotransglucosylase/hydrolase Bv-XTH1 and Bv-XTH2 promoters reveals overlapping tissue-specific and wound-inducible expression profiles. Plant Biotechnology Journal, 2, 127–139.
Faccioli, P., Ciceri, G. P., Provero, P., Stanca, A. M., Morcia, C., & Terzi, V. (2007). A combined strategy of “in silico” transcriptome analysis and web search engine optimization allows an agile identification of reference genes suitable for normalization in gene expression studies. Plant Molecular Biology, 63, 679–688.
Freialdenhoven, A., Orme, J., Lahaye, T., & Schulze-Lefert, P. (2005). Barley Rom1 reveals a potential link between race-specific and nonhost resistance responses to powdery mildew fungi. Molecular Plant–Microbe Interactions, 18, 291–299.
Freialdenhoven, A., Peterhansel, C., Kurth, J., Kreuzaler, F., & Schulze-Lefert, P. (1996). Identification of genes required for the function of non-race-specific mlo resistance to powdery mildew in barley. Plant Cell, 8, 5–14.
Fuchs, W. H. (1966). Liberation and behaviour of spores of Polymyxa betae Keskin. In M. F. Madelin (Ed.), The fungus spore (pp. 111–112). London: Butterworths Scientific Publications.
Ham, J. H., Kim, M. G., Lee, S. Y., & Mackey, D. (2007). Layered defenses underlie non-host resistance of Arabidopsis to Pseudomonas syringae pv. phaseolicola. Plant Journal, 51, 604–616.
Heath, M. C. (2000). Nonhost resistance and nonspecific plant defenses. Current Opinion in Plant Biology, 3, 315–319.
Holt, B. F., Belkhadir, Y., & Dangl, J. L. (2005). Antagonistic control of disease resistance protein stability in the plant immune system. Science, 309, 929–932.
Huitema, E., Vleeshouwers, V., Francis, D. M., & Kamoun, S. (2003). Active defence responses associated with non-host resistance of Arabidopsis thaliana to the oomycete pathogen Phytophthora infestans. Molecular Plant Pathology, 4, 487–500.
Humphry, M., Consonni, C., & Panstruga, R. (2006). mlo-based powdery mildew immunity: Silver bullet or simply non-host resistance? Molecular Plant Pathology, 7, 605–610.
Irizarry, R. A., Hobbs, B., Collin, F., Beazer-Barclay, Y. D., Antonellis, K. J., Scherf, U., & Speed, T. P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics, 4, 249–264.
Jones, J. D. G., & Dangl, J. L. (2006). The plant immune system. Nature, 444, 323–329.
Jwa, N. S., Agrawal, G. K., Tamogami, S., Yonekura, M., Han, O., Iwahashi, H., & Rakwal, R. (2006). Role of defense/stress-related marker genes, proteins and secondary metabolites in defining rice self-defense mechanisms. Plant Physiology & Biochemistry, 44, 261–273.
Kanyuka, K., Ward, E., & Adams, M. J. (2003). Polymyxa graminis and the cereal viruses it transmits: A research challenge. Molecular Plant Pathology, 4, 393–406.
Keon, J., Antoniw, J., Carzaniga, R., Deller, S., Ward, J. L., Baker, J. M., et al. (2007). Transcriptional adaptation of Mycosphaerella graminicola to programmed cell death (PCD) of its susceptible wheat host. Molecular Plant–Microbe Interactions, 20, 178–193.
Keskin, B. (1964). Polymyxa betae nsp ein parasit in den wurzeln von Beta vulgaris Tournefort besonders wahrend der jugendentwicklung der zuckerrube. Archiv Fur Mikrobiologie, 49, 348–374.
Kingsnorth, C. S., Asher, M. J. C., Keane, G. J. P., Chwarszczynska, D. M., Luterbacher, M. C., & Mutasa-Göttgens, E. S. (2003). Development of a recombinant antibody ELISA test for the detection of Polymyxa betae and its use in resistance screening. Plant Pathology, 52, 673–680.
McGrann, G. R. D., Martin, L. D., Kingsnorth, C. S., Asher, M. J. C., Adams, M. J., & Mutasa-Göttgens, E. S. (2007). Screening for genetic elements involved in the nonhost response of sugar beet to the plasmodiophorid cereal root parasite Polymyxa graminis by representational difference analysis. Journal of General Plant Pathology, 73, 260–265.
Muskett, P. R., Kahn, K., Austin, M. J., Moisan, L. J., Sadanandom, A., Shirasu, K., et al. (2002). Arabidopsis RAR1 exerts rate-limiting control of R gene-mediated defenses against multiple pathogens. Plant Cell, 14, 979–992.
Mutasa-Göttgens, E. S., Chwarszczynska, D. M., Halsey, K., & Asher, M. J. C. (2000). Specific polyclonal antibodies for the obligate plant parasite Polymyxa—A targeted recombinant DNA approach. Plant Pathology, 49, 276–287.
Peterhansel, C., Freialdenhoven, A., Kurth, J., Kolsch, R., & Schulze-Lefert, P. (1997). Interaction analyses of genes required for resistance responses to powdery mildew in barley reveal distinct pathways leading to leaf cell death. Plant Cell, 9, 1397–1409.
Rush, C. M. (2003). Ecology and epidemiology of Benyviruses and plasmodiophorid vectors. Annual Review of Phytopathology, 41, 567–592.
Shirasu, K., Lahaye, T., Tan, M. W., Zhou, F. S., Azevedo, C., & Schulze-Lefert, P. (1999). A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C-elegans. Cell, 99, 355–366.
Smyth, G. K. (2004). Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetic and Molecular Biology, 3, 3.
Thilmony, R., Underwood, W., & He, S. Y. (2006). Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157: H7. Plant Journal, 46, 34–53.
Thordal-Christensen, H. (2003). Fresh insights into processes of nonhost resistance. Current Opinion in Plant Biology, 6, 351–357.
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology, 3, 0034.0031–0034.0011.
Ward, E., & Adams, M. J. (1998). Analysis of ribosomal DNA sequences of Polymyxa species and related fungi and the development of genus- and species-specific PCR primers. Mycological Research, 102, 965–974.
Zimmerli, L., Stein, M., Lipka, V., Schulze-Lefert, P., & Somerville, S. (2004). Host and non-host pathogens elicit different jasmonate/ethylene responses in Arabidopsis. Plant Journal, 40, 633–646.
Acknowledgements
We thank Paul Schulze-Lefert, Max-Planck Institute for Plant Breeding Research, Cologne, Germany, for seeds of the barley rar1, ror1 and ror2 mutants and their respective wild-types. We also thank Simon Hodge and Aiming Qi for statistical analysis of the qRT-PCR data and Kim Hammond-Kosack, Jason Rudd and Kostya Kanyuka for strategic contributions. Contract research services were provided by Geneservice Ltd. (www.geneservice.co.uk).This project was sponsored by the Biotechnology and Biological Sciences Research Council (BBSRC) through the Sustainable Arable LINK Programme.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
McGrann, G.R.D., Townsend, B.J., Antoniw, J.F. et al. Barley elicits a similar early basal defence response during host and non-host interactions with Polymyxa root parasites. Eur J Plant Pathol 123, 5–15 (2009). https://doi.org/10.1007/s10658-008-9332-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-008-9332-z