Abstract
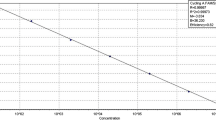
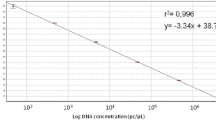
A specific primer couple (E3–E4) amplifying a single DNA fragment of 111 bp from plasmid pEA29 was designed to identify, detect and quantify Erwinia amylovora by real-time Scorpion-PCR. Specificity of primers and probe was assessed both by means of BLAST analyses and by using genomic DNA from a large number of E. amylovora isolates and other bacteria. In Scorpion-PCR, the limit of detection was of 1 pg of total DNA and a high correlation (r = 0.999) was achieved between target DNA quantity and cycle threshold (Ct). Combining two sequential amplifications with conventional reported primers (PEANT1–PEANT2) and Scorpion primers (E3 Scorpion-E4) the detection limit was of 1 fg (nested Scorpion-PCR). Using serial dilution of the bacterial suspensions the limit of detection was 3.2 × 104 CFU ml−1 in Scorpion-PCR and 2.8 × 102 CFU ml−1 in nested Scorpion-PCR. Real-time PCR combined with effective procedures for DNA extraction enabled the detection and the quantification of the epiphytic population of E. amylovora in the washings of flowers and leaves of artificially inoculated pear. A significant correlation (r = 0.92) was achieved between pathogen CFU on semi-selective media and the corresponding target DNA concentration evaluated by real-time PCR.







Similar content being viewed by others
References
Bereswill, S., Bugert, P., Bruchmuller, I., & Geider, K. (1995). Identification of the fire blight pathogen, Erwinia amylovora, by PCR assays with chromosomal DNA. Applied and Environmental Microbiology, 61, 2636–2642.
Bereswill, S., Pahl, P., Bellerman, P., Zeller, W., & Geider, K. (1992). Sensitive and species-specific detection of Erwinia amylovora by PCR analysis. Applied and Environmental Microbiology, 58, 3522–3526.
Bonn, W. G., & van der Zwet, T. (2000). Distribution and economic importance of Fire Blight. In J. L. Vanneste (Ed.), Fire Blight: The disease and its causative agent Erwinia amylovora (pp. 235–251). Wallingford, UK: CABI Publishing.
Crosse, J. E., & Goodman, R. N. (1973). A selective medium for and a definitive colony characteristic of Erwinia amylovora. Phytopathology, 63, 1425–1426.
Falkenstein, H., Bellemann, P., Walter, S., Zeller, W., & Geider, K. (1988). Identification of Erwinia amylovora, the fireblight pathogen, by colony hybridization with plasmid pEA29. Applied and Environmental Microbiology, 54, 2798–2802.
Ginzinger, D. G. (2002). Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Experimental Hematology, 30, 503–512.
Hayden, K. J., Rizzo, D., Tse, J., & Garbelotto, M. (2004). Detection and quantification of Phytophthora ramorum from California forests using a real-time polymerase chain reaction assay. Phytopathology, 94, 1075–1083.
Ippolito, A., Schena, L., Nigro, F., Soleti Ligorio, V., & Yaseen, T. (2004). Real-time detection of Phytophthora nicotianae and P. citrophthora in citrus roots and soil. European Journal of Plant Pathology, 110, 833–843.
Johnson, K. B., & Stockwell, V. O. (1998). Management of fire blight: A case study in microbial ecology. Annual Review of Phytopathology, 36, 227–248.
Llop, P., Bonaterra, A., Penalver, J., & Lopez, M. M. (2000). Development of highly sensitive nested-PCR procedure using a single closed tube for detection of Erwinia amylovora in asymptomatic plant material. Applied and Environmental Microbiology, 66, 2071–2078.
McGhee, G. C., & Jones, A. L. (2000). Complete nucleotide sequence of ubiquitous plasmid pEA29 from Erwinia amylovora strain Ea88: gene organization and intraspecies variation. Applied and Environmental Microbiology, 66, 4897–4907.
McManus, P. S., & Jones, A. L. (1995). Detection of Erwinia amylovora by nested PCR and PCR-dot-blot and reverse blot hybridizations. Phytopathology, 85, 618–623.
Momol, M., Norelli, J., Piccioni, D., Momol, E. A., Gustafson, H. L., Cummins, J. N., & Aldwinckle, H. S. (1998). Internal movement of Erwinia amylovora through symptomless apple scion tissues into the rootstock. Plant Disease, 82, 646–650.
Salm, H., & Geider, K. (2004). Real-time PCR for detection and quantification of Erwinia amylovora, the causal agent of fireblight. Plant Pathology, 53, 602–610.
Schaad, N. W., Frederick, R. D., Shaw, J., Schneider, W. L., Hickson, R., Petrillo, M. D., & Luster, D. G. (2003). Advances in molecular-based diagnostics in meeting crop biosecurity and phytosanitary issues. Annual Review of Phytopathology, 41, 305–324.
Schena, L., & Ippolito, A. (2003). Rapid, and sensitive detection of Rosellinia necatrix in roots and soils by real-time Scorpion-PCR. Journal of Plant Pathology, 85, 15–25.
Schena, L., Nigro, F., & Ippolito, A (2002). Identification and detection of Rosellinia necatrix by conventional and real-time Scorpion-PCR. European Journal of Plant Pathology, 108, 355–366.
Schena, L., Nigro, F., Ippolito, A., & Gallitelli, D. (2004). Real-time quantitative PCR: a new technology to detect and study phytopathogenic and antagonistic fungi. European Journal of Plant Pathology, 110, 893–908.
Solinas, A., Brown, L. J., McKeen, C., Mellor, J. M., Nicol, J. T. G., Thelwell, N., & Brown, T. (2001). Duplex Scorpion primers in SNP analysis and FRET applications. Nucleic Acids Research, 29, 96–104.
Taylor, R. K., Guilford, P. J., Clark, R. G., Hale, C. N., & Forster, R. L. S. (2001). Detection of Erwinia amylovora in plant material using novel polymerase chain reaction (PCR) primers. New Zealand Journal of Crop and Horticultural Science, 29, 35–43.
Thelwell, N., Millington, S., Solinas, A., Booth, J., & Brown, T. (2000). Mode of action and application of Scorpion primers to mutation detection. Nucleic Acids Research, 28, 3752–3761.
Thomson, S. V. (1986). The role of the stigma in fire blight infections. Phytopathology, 76, 476–482.
Thomson, S. V. (2000). Epidemiology of fire blight. In J. L. Vanneste (Ed.), Fire Blight: the disease and its causative agent, Erwinia amylovora (pp. 9–36). Wallingford, UK: CABI Publishing.
van der Zwet, T. (2002) Present worldwide distribution of fire blight. Acta Horticulturae, 590, 33–34.
Vanneste, J. L. (1995). Erwinia amylovora. In U. S. Singh, R. P. Singh, & K. Kohomoto (Eds.), Pathogenesis and host specificity in plant diseases: Histopathological, biochemical, genetic and molecolar bases, (Vol. 1) (33 pp). Oxford and London: Pergammon Press.
Vanneste, J. L. (2000). Fire blight: the disease and its causative agent, Erwinia amylovora. CABI Publishing, Wallingford, UK.
Whitcombe, D., Theaker, J., Guy, S. P., Brown, T., Little, S. (1999). Detection of PCR products using self-probing amplicons and fluorescence. Nature Biotechnology, 17, 804–807.
Acknowledgements
This work was supported by a grant from the University of Bari: ‘Epidemiology and genetics of plant-pathogen microrganisms’. We thank J.D. Janse Jr. at Department of Bacteriology, Plant Protection Service, Wageningen, The Netherlands, C. Bazzi at Department of Agroenvironmental Sciences and Technologies, University of Bologna, Italy, M. Scortichini at Fruit Tree Research Institute, Rome, Italy, and L. Corazza at Plant Pathology Research Institute, Rome, Italy, for kindly supplying bacterial isolates.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
De Bellis, P., Schena, L. & Cariddi, C. Real-time Scorpion-PCR detection and quantification of Erwinia amylovora on pear leaves and flowers. Eur J Plant Pathol 118, 11–22 (2007). https://doi.org/10.1007/s10658-006-9078-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-006-9078-4