Abstract
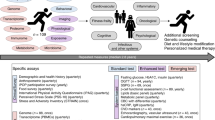
The 10 K is a large-scale prospective longitudinal cohort and biobank that was established in Israel. The primary aims of the study include development of prediction models for disease onset and progression and identification of novel molecular markers with a diagnostic, prognostic and therapeutic value. The recruitment was initiated in 2018 and is expected to complete in 2021. Between 28/01/2019 and 13/12/2020, 4,629 from the expected 10,000 participants were recruited (46 %). Follow-up visits are scheduled every year for a total of 25 years. The cohort includes individuals between the ages of 40 and 70 years. Predefined medical conditions were determined as exclusions. Information collected at baseline includes medical history, lifestyle and nutritional habits, vital signs, anthropometrics, blood tests results, Electrocardiography, Ankle–brachial pressure index (ABI), liver US and Dual-energy X-ray absorptiometry (DXA) tests. Molecular profiling includes transcriptome, proteome, gut and oral microbiome, metabolome and immune system profiling. Continuous measurements include glucose levels using a continuous glucose monitoring device for 2 weeks and sleep monitoring by a home sleep apnea test device for 3 nights. Blood and stool samples are collected and stored at − 80 °C in a storage facility for future research. Linkage is being established with national disease registries.


Similar content being viewed by others
Data Availability
Access to the 10 K cohort is currently not available online. Potential collaborators are encouraged to contact the Principal Investigator by e-mail (eran.segal@weizmann.ac.il) for further information.
References
Diabetes Prevention Program Research Group. Knowler WC, Fowler SE, Hamman RF, Christophi CA, Hoffman HJ, et al. 10-year follow-up of diabetes incidence and weight loss in the Diabetes Prevention Program Outcomes Study. Lancet. 2009;374:1677–86.
Knoops KTB, de Groot LCPGM, Kromhout D, Perrin A-E, Moreiras-Varela O, Menotti A, et al. Mediterranean diet, lifestyle factors, and 10-year mortality in elderly European men and women: the HALE project. JAMA. 2004;292:1433–9.
Shilo S, Rossman H, Segal E. Axes of a revolution: challenges and promises of big data in healthcare. Nat Med. 2020;26:29–38.
Kinkorová J. Biobanks in the era of personalized medicine: objectives, challenges, and innovation: Overview. EPMA J. 2015;7:4.
Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–5.
UK Biobank: Protocol for a large-scale prospective epidemiological resource avilable at https://www.ukbiobank.ac.uk/learn-more-about-ukbiobank/about-us (accessed 23 July, 2020).
Blaser MJ. Antibiotic use and its consequences for the normal microbiome. Science. 2016;352:544–5.
Halfvarson J, Brislawn CJ, Lamendella R, Vázquez-Baeza Y, Walters WA, Bramer LM, et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol. 2017;2:17004.
Pang KP, Gourin CG, Terris DJ. A comparison of polysomnography and the WatchPAT in the diagnosis of obstructive sleep apnea. Otolaryngol Head Neck Surg. 2007;137:665–8.
Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–70.
Lawlor DA, Harbord RM, Sterne JAC, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27:1133–63.
Hernán MA, Robins JM. Using big data to emulate a target trial when a randomized trial is not available. Am J Epidemiol. 2016;183:758–64.
Hernán MA, Robins JM. Estimating causal effects from epidemiological data. J Epidemiol Commun Health. 2006;60:578–86.
Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y, et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47:856–60.
Fry A, Littlejohns TJ, Sudlow C, Doherty N, Adamska L, Sprosen T, et al. Comparison of Sociodemographic and Health-Related Characteristics of UK Biobank Participants With Those of the General Population. Am J Epidemiol. 2017;186:1026–34.
Swanson JM. The UK Biobank and selection bias. Lancet. 2012;380:110.
Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A, et al. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163:1079–94.
Zeevi D, Korem T, Godneva A, Bar N, Kurilshikov A, Lotan-Pompan M, et al. Structural variation in the gut microbiome associates with host health. Nature. 2019;568:43–8.
Bar N, Korem T, Weissbrod O, Zeevi D, Rothschild D, Leviatan S, et al. A reference map of potential determinants for the human serum metabolome. Nature. 2020;588:135.
Acknowledgements
The authors wish to acknowledge all participants of the cohort and the members of the Segal lab for fruitful discussions.
Funding
E.S. is supported by the Crown Human Genome Center; Larson Charitable Foundation New Scientist Fund; Else Kroener Fresenius Foundation; White Rose International Foundation; Ben B. and Joyce E. Eisenberg Foundation; Nissenbaum Family; Marcos Pinheiro de Andrade and Vanessa Buchheim; Lady Michelle Michels; Aliza Moussaieff; and grants funded by the Minerva foundation with funding from the Federal German Ministry for Education and Research and by the European Research Council and the Israel Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All author declares that they have no conflict of interest.
Consent to participate
All participants provided written informed consent.
Ethics approval
Ethics approval was granted by the Institutional Review Board (IRB) of the Weizmann Institute of Science.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shilo, S., Bar, N., Keshet, A. et al. 10 K: a large‐scale prospective longitudinal study in Israel. Eur J Epidemiol 36, 1187–1194 (2021). https://doi.org/10.1007/s10654-021-00753-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10654-021-00753-5