Abstract
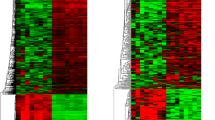
Temozolomide is a first line anti-tumor drug used for the treatment of patients with Glioblastoma multiforme (GBM). However, the drug resistance to temozolomide limits its clinical application. Therefore, novel strategies to overcome chemoresistance are desperately needed for improved treatment of human GBM. Here, we simultaneously detected, for the first time, the expression profiles of mRNAs, lncRNAs, and circRNAs in three pairs of secondary temozolomide-resistant glioblastoma (STRG) and matched primary glioblastoma tissues by microarrays. Using these data, we discovered a total of 92 mRNA, 299 lncRNAs and 53 circRNAs were altered in human glioma tissue after chemotherapy with temozolomide. The functions of differentially expressed lncRNAs, circRNAs were annotated by analysis of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG). The results showed that the highest enriched GO terms of the upregulated lncRNAs were embryonic forelimb morphogenesis (BP), extracellular space (CC), and serine-type endopeptidase activity (MF). Meanwhile, GO:0035360(BP), PRC1 complex (CC), and ubiquitin-protein transferase activity (MF) were the highest enriched GO terms targeted by downregulated lncRNAs. The NF-kappa B signaling pathway were significantly enriched in the STRG. However, circRNAs highest enriched GO term was viral process, chromosome, and protein transporter activity, respectively. KEGG pathway analysis showed that circRNAs in the network were enriched in ErbB signaling pathway. Furthermore, we also predicted the potential role of these differentially expressed ncRNAs and constructed a network of lncRNAs-mRNAs and circRNAs-miRNAs to show their interactions. After a series of bioinformatics analyses, we found that low expression of NONHSAT163779 and high expression of circ_0043949 are closely related to the chemoresistance of STRG. Our findings revealed the alteration of expression patterns of mRNAs, lncRNAs, and circRNAs in the secondary temozolomide-resistant glioblastoma for the first time. NONHSAT163779 and hsa_circ_0043949 might be potential therapeutic targets and prognostic biomarkers for the treatment of glioblastoma.




Similar content being viewed by others
References
Puchalski RB, Shah N, Miller J, Dalley R, Nomura SR, Yoon JG, Smith KA, Lankerovich M, Bertagnolli D, Bickley K, Boe AF, Brouner K, Butler S, Caldejon S, Chapin M, Datta S, Dee N, Desta T, Dolbeare T, Dotson N, Ebbert A, Feng D, Feng X, Fisher M, Gee G, Goldy J, Gourley L, Gregor BW, Gu G, Hejazinia N, Hohmann J, Hothi P, Howard R, Joines K, Kriedberg A, Kuan L, Lau C, Lee F, Lee H, Lemon T, Long F, Mastan N, Mott E, Murthy C, Ngo K, Olson E, Reding M, Riley Z, Rosen D, Sandman D, Shapovalova N, Slaughterbeck CR, Sodt A, Stockdale G, Szafer A, Wakeman W, Wohnoutka PE, White SJ, Marsh D, Rostomily RC, Ng L, Dang C, Jones A, Keogh B, Gittleman HR, Barnholtz-Sloan JS, Cimino PJ, Uppin MS, Keene CD, Farrokhi FR, Lathia JD, Berens ME, Iavarone A, Bernard A, Lein E, Phillips JW, Rostad SW, Cobbs C, Hawrylycz MJ, Foltz GD (2018) An anatomic transcriptional atlas of human glioblastoma[J]. SCIENCE 360(6389):660–663
Northcott PA (2017) Cancer: keeping it real to kill glioblastoma[J]. Nature 547(7663):291–292
Hilf N, Kuttruff-Coqui S, Frenzel K, Bukur V, Stevanović S, Gouttefangeas C, Platten M, Tabatabai G, Dutoit V, van der Burg S, Thor Straten P, Martínez-Ricarte F, Ponsati B, Okada H, Lassen U, Admon A, Ottensmeier CH, Ulges A, Kreiter S, von Deimling A, Skardelly M, Migliorini D, Kroep JR, Idorn M, Rodon J, Piró J, Poulsen HS, Shraibman B, McCann K, Mendrzyk R, Löwer M, Stieglbauer M, Britten CM, Capper D, Welters MJP, Sahuquillo J, Kiesel K, Derhovanessian E, Rusch E, Bunse L, Song C, Heesch S, Wagner C, Kemmer-Brück A, Ludwig J, Castle JC, Schoor O, Tadmor AD, Green E, Fritsche J, Meyer M, Pawlowski N, Dorner S, Hoffgaard F, Rössler B, Maurer D, Weinschenk T, Reinhardt C, Huber C, Rammensee HG, Singh-Jasuja H, Sahin U, Dietrich PY, Wick W (2019) Actively personalized vaccination trial for newly diagnosed glioblastoma[J]. Nature 565(7738):240–245
Desjardins A, Gromeier M, Herndon JE 2nd et al (2018) Recurrent Glioblastoma treated with recombinant poliovirus[J]. N Engl J Med 379(2):150–161
Banelli B, Forlani A, Allemanni G et al (2017) MicroRNA in Glioblastoma: an overview[J]. Int J Genomics 2017:7639084
Lei B, Yu L, Jung TA, Deng Y, Xiang W, Liu Y, Qi S (2018) Prospective series of nine Long noncoding RNAs associated with survival of patients with Glioblastoma[J]. J Neurol Surg A Cent Eur Neurosurg 79(6):471–478
Gao WZ, Guo LM, Xu TQ, Yin YH, Jia F (2018) Identification of a multidimensional transcriptome signature for survival prediction of postoperative glioblastoma multiforme patients[J]. J Transl Med 16(1):368
Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T (2018) Emerging roles of long non-coding RNA in cancer[J]. Cancer Sci 109(7):2093–2100
Chen L, Dzakah EE, Shan G (2018) Targetable long non-coding RNAs in cancer treatments[J]. Cancer Lett 418:119–124
Arun G, Diermeier SD, Spector DL (2018) Therapeutic targeting of Long non-coding RNAs in Cancer[J]. Trends Mol Med 24(3):257–277
Murugan AK, Munirajan AK, Alzahrani AS (2018) Long noncoding RNAs: emerging players in thyroid cancer pathogenesis[J]. Endocr Relat Cancer 25(2):R59–R82
Zhao J, Du P, Cui P et al (2018) LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer[J]. Oncogene 37(30):4094–4109
Wang SH, Zhang MD, Wu XC, Weng MZ, Zhou D, Quan ZW (2016) Overexpression of LncRNA-ROR predicts a poor outcome in gallbladder cancer patients and promotes the tumor cells proliferation, migration, and invasion[J]. Tumour Biol 37(9):12867–12875
Chen F, Li Z, Deng C, Yan H (2019) Integrated analysis identifying new lncRNA markers revealed in ceRNA network for tumor recurrence in papillary thyroid carcinoma and build of nomogram[J]. J Cell Biochem 120(12):19673–19683
Huang MJ, Zhao JY, Xu JJ et al (2019) lncRNA ADAMTS9-AS2 controls human Mesenchymal stem cell Chondrogenic differentiation and functions AS a ceRNA[J]. Mol Ther Nucleic Acids 18:533–545
Zang J, Lu D, Xu A (2020) The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function[J]. J Neurosci Res 98(1):87–97
Verduci L, Strano S, Yarden Y, Blandino G (2019) The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment[J]. Mol Oncol 13(4):669–680
Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL (2018) CircRNA: a novel type of biomarker for cancer[J]. Breast Cancer 25(1):1–7
Yin Y, Long J, He Q, Li Y, Liao Y, He P, Zhu W (2019) Emerging roles of circRNA in formation and progression of cancer[J]. J Cancer 10(21):5015–5021
Wang ZL, Wang C, Liu W et al (2019) Emerging roles of the long non-coding RNA 01296/microRNA-143-3p/MSI2 axis in development of thyroid cancer[J]. Biosci Rep
Li Q, Jia H, Li H, Dong C, Wang Y, Zou Z (2016) LncRNA and mRNA expression profiles of glioblastoma multiforme (GBM) reveal the potential roles of lncRNAs in GBM pathogenesis[J]. Tumour Biol 37(11):14537–14552
Bissar-Tadmouri N, Donahue WL, Al-Gazali L et al (2014) X chromosome exome sequencing reveals a novel ALG13 mutation in a nonsyndromic intellectual disability family with multiple affected male siblings[J]. Am J Med Genet A 164A(1):164–169
Hamici S, Bastaki F, Khalifa M (2017) Exome sequence identified a c.320A > G ALG13 variant in a female with infantile epileptic encephalopathy with normal glycosylation and random X inactivation: review of the literature[J]. European Journal of Medical Genetics 60(10):541–547
Gao P, Wang F, Huo J, Wan D, Zhang J, Niu J, Wu J, Yu B, Sun T (2019) ALG13 deficiency associated with increased seizure susceptibility and severity[J]. NEUROSCIENCE 409:204–221
Zhang F, Cui Y (2019) Dysregulation of DNA methylation patterns may identify patients with breast cancer resistant to endocrine therapy: a predictive classifier based on differentially methylated regions[J]. Oncol Lett 18(2):1287–1303
De Antonellis P, Carotenuto M, Vandenbussche J et al (2014) Early targets of miR-34a in neuroblastoma[J]. Mol Cell Proteomics 13(8):2114–2131
Luo J, Liu H, Luan S, Li Z (2019) Guidance of circular RNAs to proteins' behavior as binding partners[J]. Cell Mol Life Sci 76(21):4233–4243
Zaiou M (2019) Circular RNAs in hypertension: challenges and clinical promise[J]. Hypertens Res 42(11):1653–1663
Wang KW, Dong M (2019) Role of circular RNAs in gastric cancer: recent advances and prospects[J]. World J Gastrointest Oncol 11(6):459–469
Zhang QY, Men CJ, Ding XW (2019) Upregulation of microRNA-140-3p inhibits epithelial-mesenchymal transition, invasion, and metastasis of hepatocellular carcinoma through inactivation of the MAPK signaling pathway by targeting GRN[J]. J Cell Biochem 120(9):14885–14898
Zhao K, Li X, Chen X, Zhu Q, Yin F, Ruan Q, Xia J, Niu Z (2019) Inhibition of miR-140-3p or miR-155-5p by antagomir treatment sensitize chordoma cells to chemotherapy drug treatment by increasing PTEN expression[J]. Eur J Pharmacol 854:298–306
Ho KH, Cheng CH, Chou CM et al (2019) miR-140 targeting CTSB signaling suppresses the mesenchymal transition and enhances temozolomide cytotoxicity in glioblastoma multiforme[J]. Pharmacol Res 147:104390
Funding
The work was supported by the National Natural Science Foundation of China (U1504819), the Foundation for Innovative Research Groups of the First Affiliated Hospital of Zhengzhou University (TD2011023) and the Scientific and Technological Project of Henan Province (112102310173).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
Author Chengbin Zhao declares that he has no conflict of interest. Author Yuyuan Gao declares that he has no conflict of interest. Author Ruiming Guo declares that he has no conflict of interest. Author Hongwei Li declares that he has no conflict of interest. Author Bo Yang declares that he has no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent (optional)
For this type of study, Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Zhao, C., Gao, Y., Guo, R. et al. Microarray expression profiles and bioinformatics analysis of mRNAs, lncRNAs, and circRNAs in the secondary temozolomide-resistant glioblastoma. Invest New Drugs 38, 1227–1235 (2020). https://doi.org/10.1007/s10637-019-00884-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10637-019-00884-3