Summary
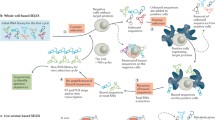
Aptamers are short single-stranded DNA or RNA oligonucleotides that are capable of binding small molecules, proteins, or nucleotides with high specificity. They show a stable conformation and high binding affinity for their target molecules. There are numerous applications for aptamers in biotechnology, molecular diagnostics and targeted therapy of diseases. Their production is cheap, and they generally display lower immunogenicity than monoclonal antibodies. In the present review, we give an introduction to the preparation of aptamers and provide examples for their use in biotechnology, diagnostics and therapy of diseases.

Similar content being viewed by others
References
Förstermann U, Kleinert H (2009) Medizinische Gentechnologie und Gentherapie. In: Aktories K, Förstermann U, Hofmann FB, Starke K (eds) Allgemeine und spezielle Pharmakologie und Toxikologie. Elsevier GmbH, 10. Auflage, München, pp 24–35
Mayer G (2009) The chemical biology of aptamers. Angew Chem Int Ed 48:2672–2689
Rehm H, Letzel T (2010) Der Experimentator – Proteinbiochemie/ Proteomics, 6th edn. Auflage, Heidelberg, p 314
Collett JR, Cho EJ, Ellington AD (2005) Production and processing of aptamer microarrays. Methods 37:4–15
Hall DA, Ptacek J, Snyder M (2007) Protein microarray technology. Mech Ageing Dev 128:161–167
Lee JH, Yigit MV, Mazumdar D, Lu Y (2010) Molecular diagnostic and drug delivery agents based on aptamer-nanomaterial conjugates. Adv Drug Deliv Rev 62:592–605
Langer R (1998) Drug delivery and targeting. Nature 392(6679):5–10
Wilson C, Szostak JW (1998) Isolation of a fluorophore-specific DNA aptamer with weak redox activity. Chem Biol 5:609–617
Farokhzad OC, Cheng J, Teply BA, Sherifi I, Jon S, Kantoff PW, Richie JP, Langer R (2006) Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc Natl Acad Sci U S A 103:6315–6320
Liss M, Petersen B, Wolf H, Prohaska E (2002) An aptamer-based quartz crystal protein biosensor. Anal Chem 74:4488–4495
Kanwar JR, Roy K, Kanwar RK (2011) Chimeric aptamers in cancer cell-targeted drug delivery. Crit Rev Biochem Mol Biol 46:459–477
Thompson KM, Syrett HA, Knudsen SM, Ellington AD (2002) Group I aptazymes as genetic regulatory switches. BMC Biotechnol 2:21
Isaacs FJ, Dwyer DJ, Ding C, Pervouchine DD, Cantor CR, Collins JJ (2004) Engineered riboregulators enable post-transcriptional control of gene expression. Nat Biotechnol 22:841–847
Suess B, Fink B, Berens C, Stentz R, Hillen W (2004) A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Res 32:1610–1614
Laserson U, Gan HH, Schlick T (2005) Predicting candidate genomic sequences that correspond to synthetic functional RNA motifs. Nucleic Acids Res 33:6057–6069
Lee JH, Wernette DP, Yigit MV, Liu J, Wang Z, Lu Y (2008) Site-specific control of distances between gold nanoparticles using phosphorothioate anchors on DNA and a short bifunctional molecular fastener. Angew Chem Int Ed 46:9006–9010
Ulrich H, Wrenger C (2009) Disease-specific biomarker discovery by aptamers. Cytometry A 75:727–733
Famulok M, Hartig JS, Mayer G (2007) Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem Rev 107:3715–3743
Dassie JP, Liu XY, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR, Meyerholz DK, McCaffrey AP, McNamara JO 2nd, Giangrande PH (2009) Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat Biotechnol 27:839–849
Gold L, Polisky B, Uhlenbeck O, Yarus M (1995) Diversity of oligonucleotide functions. Annu Rev Biochem 64:763–797
Burmeister PE, Lewis SD, Silva RF, Preiss JR, Horwitz LR, Pendergrast PS, McCauley TG, Kurz JC, Epstein DM, Wilson C, Keefe AD (2005) Direct in vitro selection of a 2′-O-methyl aptamer to VEGF. Chem Biol 12:25–33
Dausse E, Da Rocha GS, Toulmé JJ (2009) Aptamers: a new class of oligonucleotides in the drug discovery pipeline? Curr Opin Pharmacol 9:602–607
Thiel KW, Giangrande PH (2009) Therapeutic applications of DNA and RNA aptamers. Oligonucleotides 19:209–222
Leva S, Lichte A, Burmeister J, Muhn P, Jahnke B, Fesser D, Jeannette E, Burgstaller P, Klussmann S (2002) GnRH binding RNA and DNA spiegelmers: a novel approach toward GnRH antagonism. Chem Biol 9:351–359
Frauendorf C, Hausch F, Röhl I, Lichte A, Vonhoff S, Klussmann S (2003) Internal 32P-labeling of L-deoxyoligonucleotides. Nucleic Acids Res 31:e34
Vater A (2004) Entwicklung eines Verfahrens zur Identifizierung kurzer hochaffiner RNA-Oligonukleotide am Beispiel von CGRP-antagonisierenden Spiegelmeren. Tenea Verlag, Berlin, pp 13–15
Jarosch F (2005) Automatisierte Verfahren zur Selektion kurzer RNA- und DNA-Spiegelmere. Tenea Verlag, Berlin, pp 12–16
Grisanti S (2008) Pegabtanib Macugen®. In: Bartz-Schmidt KU, Ziemssen F (eds) Intravitreale Pharmakotherapie-Moderne Medikamente und ihre Anwendung am Auge. Schattauer GmbH, Stuttgart, p 75
Ellington AD, Szostak JW (1992) Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature 355:850–852
Wlotzka B, Leva S, Eschgfäller B, Burmeister J, Kleinjung F, Kaduk C, Muhn P, Hess-Stumpp H, Klussmann S (2002) In vivo properties of an Anti-GnRH Spiegelmer: an example of an oligonucleotide-based therapeutic substance class. Proc Natl Acad Sci U S A 99:8898–8902
Habermehl GG, Krebs HC, Hammann PE, Ternes W (2008) Naturstoffchemie, 3rd edn. Springer, Berlin, p 458
Eulberg D, Buchner K, Maasch C, Klussmann S (2005) Development of an automated in vitro selection protocol to obtain RNA-based aptamers: identification of a biostable substance P antagonist. Nucleic Acids Res 33:e45
Gold L, Walker JJ, Wilcox SK, Williams S (2012) Advances in human proteomics at high scale with the SOMAscan proteomics platform. Nat Biotechnol 29:543–539
Sinha J, Reyes SJ, Gallivan JP (2010) Reprogramming bacteria to seek and destroy an herbicide. Nat Chem Biol 6:464–470
Ilyas A, Asghar W, Allen PB, Duhon H, Ellington AD, Igbal SM (2012) Electrical detection of cancer biomarkers using aptamers with nanogap break junctions. Nanotechnology 23:275502
Heintz A, Reinhardt GA (1996) Chemie und Umwelt, 4th edn. Aktualisierte und erweiterte Auflage, Braunschweig, p 217
Schachat AP (2005) New treatments for age-related macular degeneration. Ophthalmology 112:531–532
Lee JH, Canny MD, De Erkenez A, Krilleke D, Ng YS, Shima DT, Pardi A, Jucker F (2005) A therapeutic aptamer inhibits angiogenesis by specifically targeting the heparin binding domain of VEGF165. Proc Natl Acad Sci USA 102:18902–18907
Clark DP, Pazdernik NJ (2009) Molekulare Biotechnologie-Grundlagen und Anwendungen. Spektrum Akademischer Verlag, Heidelberg, p 472
Do DV, Haller JA, Adamis AP, Striata C, Nguyen QD, Shah SM, Joussen AM (2008) Anti-VEGF therapy as an emerging treatment for diabetic retinopathy. In: Duh, E. (Eds.) Diabetic retinopathy. Human Press, Springer Verlag, 406–407
Ng EWM, Shima DT, Calias P, Cunningham ET Jr, Guyer DR, Adamis AP (2006) Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov 5:123–132
Moshfeghi AA, Puliafito CA (2005) Pegaptanib sodium for the treatment of neovascular age-related macular degeneration. Exp Opin Investig Drugs 14:671–682
Zhou J, Rossi JJ (2010) Aptamer-targeted cell-specific RNA interference. Silence 1:4
Zhou J, Bobbin ML, Burnett JC, Rossi JJ (2012) Current progress of RNA aptamer-based therapeutics. Front Genet 3:234
Lupold SE, Hicke BJ, Lin Y, Coffey DS (2002) Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res 62:4029–4033
Chu TC, Marks JW 3rd, Lavery LA, Faulkner S, Rosenblum MG, Ellington AD (2006) Aptamer: toxin conjugates that specifically target prostate tumor cells. Cancer Res 66:5989–5992
Stecker JR, Savage AA, Bruno JG, Garcia DM, Koke JR (2012) Dynamics and visualization of MCF7 adenocarcinoma cell death by aptamer-C1q-mediated membrane attack. Nucleic Acid Ther 22:275–282
Liu Z, Duan JH, Song YM, Ma J, Wang FD, Lu X, Yang XD (2012) Novel HER2 aptamer selectively delivers cytotoxic drug to HER2-positive breast cancer cells in vitro. J Transl Med 10:148
Cao ZH, Tong R, Mishra A, Xu W, Wong GC, Cheng J, Lu Y (2009) Reversible cell-specific drug delivery with aptamer-functionalized liposomes. Angew Chem Int Ed 48:6494–6498
Kanwar JR, Roy K, Kanwar RK (2011) Chimeric aptamers in cancer cell-targeted drug delivery. Crit Rev Biochem Mol Biol 46:459–477
Aulbert E, Disselhoff W, Sörje H, Schulz E, Gericke D (1980) Lysosomal accumulation of 67Ga-transferrin in malignant tumors in relation to their growth rate. Eur J Cancer 16:1217–1232
McNamara JO, Kolonias D, Pastor F, Mittler RS, Chen L, Giangrande PH, Sullenger B, Gilboa E (2008) Multivalent 4-1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice. J Clin Invest 118:376–386
Gopinath SC (2008) Anti-coagulant aptamers. Thromb Res 122:838–847
Dobrovolsky AB, Titaeva EV, Khaspekova SG, Spiridonova VA, Kopylov AM, Mazurov AV (2009) Inhibition of thrombin activity with DNA-aptamers. Bull Exp Biol Med 148:33–36
Rusconi CP, Scardino E, Layzer J, Pitoc GA, Ortel TL, Monroe D, Sullenger BA (2002) RNA aptamers as reversible antagonists of coagulation factor IXa. Nature 419:90–94
Hwang B, Cho JS, Yeo HJ, Kim JH, Chung KM, Han K, Jang SK, Lee SW (2004) Isolation of specific and high-affinity RNA aptamers against NS3 helicase domain of hepatitis C virus. RNA 10:1277–1290
Nishikawa F, Funaji K, Fukuda K, Nishikawa S (2004) In vitro selection of RNA aptamers against the HCVNS3 helicase domain. Oligonucleotides 14:114–129
Fukuda K, Umehara T, Sekiya S, Kunio K, Hasegawa T, Nishikawa S (2004) An RNA ligand inhibits hepatitis C virus NS3 protease and helicase activities. Biochem Biophys Res Commun 325:670–675
Zhan LS, Zhuo HL, Wang HZ, Peng JC, Wang QL. Screening and characterization of aptamers of hepatitis C virus NS3 helicase. Prog Biochem Biophys 32: 245–250
Hwang B, Lee SW (2005) Analysis of in vivo interaction of HCVNS3 protein and specific RNA aptamer with yeast three-hybrid system. J Microbiol Biotechnol 15:660–664
Romero-Lopez C, Barroso-del Jesus A, Puerta-Fernandez E, Berzal-Herranz A (2005) Interfering with hepatitis C virus IRES activity using RNA molecules identified by a novel in vitro selection method. Biol Chem 386:183–190
Kikuchi K, Umehara T, Fukuda K, Kuno A, Hasegawa T, Nishikawa S (2005) A hepatitis C virus (HCV) internal ribosome entry site (IRES) domain III-IV-targeted aptamer inhibits translation by binding to an apical loop of domain IIId. Nucleic Acids Res 33:683–692
Bellecave P, Andreola ML, Ventura M, Tarrago-Litvak L, Litvak S, Astier-Gin T (2003) Selection of DNA aptamers that bind the RNA-dependent RNA polymerase of hepatitis C virus and inhibit viral RNA synthesis in vitro. Oligonucleotides 13:455–463
Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249:505–510
Bunka DH, Stockley PG (2006) Aptamers come of age - at last. Nat Rev Microbiol 4:588–596
Held DM, Kissel JD, Patterson JT, Nickens DG, Burke DH (2006) HIV-1 inactivation by nucleic acid aptamers. Front Biosci 11:89–112
Zhou J, Li H, Li S, Zaia J, Rossi JJ (2008) Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol Ther 16:1481–1489
Wang J, Jiang H, Liu F (2000) In vitro selection of novel RNA ligands that bind human cytomegalovirus and block viral infection. RNA 6:571–583
Ylera F, Lurz R, Erdmann VA, Furste JP (2002) Selection of RNA aptamers to the Alzheimer’s disease amyloid peptide. Biochem Biophys Res Commun 290:1583–1588
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kadioglu, O., Malczyk, A.H., Greten, H.J. et al. Aptamers as a novel tool for diagnostics and therapy. Invest New Drugs 33, 513–520 (2015). https://doi.org/10.1007/s10637-015-0213-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10637-015-0213-y