Abstract
Background
Heparanase (HPSE) is considered to play an important role in the occurrence, development and carcinogenesis of ulcerative colitis (UC). There are no reports about the detection of HPSE mRNA in feces to predict UC activity and cancerization risk.
Aims
To explore the feasibility and effectiveness of fecal epithelial HPSE mRNA in monitoring patients’ UC activity and predicting cancer risk.
Methods
The clinical part of the study enrolled 20 patients with UC and 20 controls. Meanwhile, a UC-induced carcinogenesis mouse model was established using a combination treatment of dimethylhydrazine and dextran sulfate sodium. Tissue expression of HPSE protein was detected by immunohistochemistry. RT-qPCR was used to detect the expression of HPSE mRNA in colonic mucosa and feces.
Results
In the human study, the relative expressions of HPSE mRNA in colonic mucosa and feces were positively correlated with the Mayo score (P < 0.05), and with a significant correlation between feces and colonic mucosa (P < 0.05). In the mouse model, the relative expressions of HPSE mRNA in colonic mucosa and feces in the ulcerative colitis-associated colorectal cancer group was significantly higher than that of the UC group and the normal control group (P < 0.05), and with a significant correlation between feces and colonic mucosa (P < 0.05).
Conclusions
The relative level of HPSE mRNA was positively correlated with UC activity and cancerization. The relative level of HPSE mRNA in feces was correlated with that in colonic mucosa. The detection of HPSE mRNA in feces can be used as a new marker for disease monitoring and cancer risk prediction of UC.
Similar content being viewed by others
Introduction
Over the last decade, ulcerative colitis (UC) has become one of the common diseases in the digestive system and the main causes of chronic diarrhea in China. Studies have suggested that UC is prone to chronic delay and has a certain risk of cancerization. Clinical evaluation of UC mainly includes the assessments of clinical types, progression of concurrent symptoms and its significant association with cancer. At present, UC condition evaluation primarily relies on colonoscopy [1, 2], which is known as somewhat a painful procedure that in turn may lead to anxiety and depression in patients. So far, there are no ideal laboratory evaluation indexes for UC [3].
Erythrocyte sedimentation rate (ESR) and C-reactive protein (CRP) are serological indexes, which tend to increase in many pathologic conditions; yet, they have low specificity (which means that the test is more likely to be falsely positive). Currently, the Mayo score is considered the most reliable method for assessing the UC activity and monitoring patients during therapy. The Mayo score is based on calculating the bleeding, stool frequency, physician assessment and endoscopic appearance. Yet, this score may be complex to analyze and subjective [3].
Molecular-based fecal RNA detection is a new noninvasive detection with high specificity and sensitivity. This simple and easy method has become one of the main research interests in evaluation of the UC activity and prediction of the cancerization risk [4,5,6,7].
Heparanase (HPSE) is an essential enzyme that degrades extracellular matrix (ECM) and basement membrane (BM). It is involved in many physiological and pathological activities, including morphogenesis, neovascularization, inflammation and tumorigenesis [8]. HPSE protein and its mRNA are highly expressed in early colon tumors [9,10,11]. Recently, scientists have been trying to explore the relationship between HPSE and inflammatory responses. HPSE has an important role in the process of infectious inflammation and autoimmune inflammation. In the process of chronic inflammation of the colon, such as UC, HPSE can degrade the heparan sulfate (HS) chain, destroy the integrity of ECM and BM, and in turn, facilitate the directional migration of inflammatory cells and release bioactive factors on ECM and cell surface [12]. At the same time, HPSE can mediate macrophages to release TNFa, IL1, IL6 and other cytokines, and further aggravate intestinal inflammation, induce tumorigenesis and promote angiogenesis [13,14,15,16]. However, so far, the use of HPSE mRNA in feces as a biomarker for prediction of UC activity and risk of cancer has remained unexplored. Thus, the purpose of this study was to analyze the expression of HPSE mRNA in the colonic mucosa and feces of UC patients and a UC-induced carcinogenesis mouse model. The obtained data were compared to a Mayo scoring system.
Methods
Patients
A total of 20 UC patients diagnosed by endoscopy from outpatient and inpatient departments of Gastroenterology of Zhejiang Provincial People’s Hospital between December 2017 and June 2019 were enrolled in this study. Inclusion criteria were: preliminarily diagnosed patients with UC; > 18 years old, biopsies taken and informed consent. Exclusion criteria were: a history of treatment with infliximab, non-classifiable inflammatory disease, indeterminate colitis, infectious ileocolitis, asthma, history of autoimmune diseases, primary and secondary tumors, hypertension and type 2 diabetes mellitus. The male-to-female ratio was 14:6, the average age was 38.3 (21–52) years, and the average disease duration was 22.5 (7–94) months. Thirteen patients were in the active stage and seven in the remission stage of the disease. Besides, 20 healthy outpatients with normal colonoscopy and biopsy results were selected as the normal control group (the male-to-female ratio was 11:9; the average age was 41.2 (16–69) years.
The ethical committee of Zhejiang Provincial People’s Hospital (2019KY118) approved our study. All procedures performed in studies involving human participants were in accordance with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
All subjects underwent colonoscopy, biopsy and pathological examination. Two expertized gastroenterologists reviewed all the data. The diagnosis was carried out in accordance with the diagnostic guidelines formulated by the inflammatory bowel disease discipline group of Digestive Disease Branch of the Chinese Medical Association and confirmed by histopathological biopsy. All biopsies were sent to the Pathology Department of Zhejiang Provincial People’s Hospital for treatment and were analyzed by two senior pathologists. The pathological diagnosis standard mainly refers to the consensus opinion on the diagnostic standard of inflammatory bowel disease in China formulated by the collaborative group of inflammatory bowel disease of the digestive branch of the Chinese Medical Association in 2018 [1].
Specimen collection: ① Tissue specimen: All enrolled patients were examined and biopsied by colonoscopy. At least two biopsies were collected from the most affected inflammatory sites during colonoscopy. The diagnosis was made by a pathologist according to the principle of blindness. The Truelove–Witts standard was used for pathological diagnosis. ② Fecal specimen: The fecal specimen was taken 5–7 days after colonoscopy. After collections, samples were sealed and stored at − 20 °C.
Stage of disease: The UC stage was classified into the active stage and the remission stage. Southerland disease activity index, also known as the Mayo score [17], which includes diarrhea, hematochezia and mucous membrane performance, was used to evaluate UC activity. Several evaluation indexes (Table 1) of the clinical condition were considered for scoring stages: 0–2 points for the remission stage; 3–5 points for low-grade activity; 6–10 points for moderate-grade activity; 11–12 points for severe-grade activity.
Animals
Sixty male C57BL/6 mice, 6–8 weeks old, weighing 18–20 g, were purchased from Shanghai Lingchang Biotechnology Co., Ltd. (SCXK2018-003). All the animals were housed in Plexiglas® cages with a temperature of 25 ± 1 °C, relative humidity of 45 ± 1% and a light/dark cycle of 12/12 h, and had access to sterilized food and water ad libitum. Water and food consumption was recorded daily. All animal studies (including the mice euthanasia procedures) were done in compliance with the regulations and guidelines of Zhejiang Chinese Medical University institutional animal care and conducted according to the AAALAC and the IACUC (approval no. IACUC-20180521-03; Hangzhou, China) guidelines (license no. SYXK (Zhejiang) 2018-0012).
Materials
DSS (dextran sulfate sodium, 36–50 kDa, 9011-18-1) and DMH (1,2-dimethylhydrazine, D0741) were purchased from MP Company in the USA. Anti-heparanase 1 antibody (ab85543) and HRP labeled goat anti-rabbit II antibody (ab205718) were acquired from Abcam Company in the USA. SP-9000 SPlink detection kit was purchased from Beijing Zhongshanjinqiao Biotechnology Co., Ltd. Dynabeads Epithelial Enrich (16102) was purchased from Thermo Fisher Company in the USA. CD326 (EpCAM) MicroBeads mouse (130105958) was obtained from Miltenyi Biotec in Germany. Trizol reagent (15596026), RNase inhibitor and First-Strand cDNA Synthesis Kit (k1622) were all bought from Invitrogen Company of the USA; RNA-free water, RNeasy Mini Kit (74104) and Quantity Nova SYBR Green PCR kit (208052) were purchased from Qiagen from USA.
Heparanase and GAPD primers were designed using an NCBI primer blast and synthesized by Jinweizhi of Suzhou, China. TBE and 6× loading buffer were purchased from the Biyuntian Company.
Mouse Model
The animal models were established according to a previously described approach [18], with some slight modifications. Sixty specific pathogen-free (SPF) grade male C57BL/6 mice were randomly divided into three groups (20 mice/group): the normal control group, the UC group and the UCACRC group. UC was induced by drinking 3.0% (wt/vol) DSS in drinking water for 5 day and 0.5% (wt/vol) DSS in drinking water for 2 days. This feeding process was repeated three times until one of the following symptoms appeared: semi-dilute stool, diarrhea, occult blood in stool and bloody stool on naked eye examination. In the UCACRC group, mice were given 3.0% (wt/vol) DSS in drinking water for 1 week and then intraperitoneal injection of 20 mg/kg dimethylhydrazine (1,2-dimethylhydrazine, DMH) once, and normal saline (NS) as drinking water for 2 weeks. This process was repeated four times. After the model was established, mice in each group were individually housed for 48 h to collect feces; fecal samples were stored at − 20 °C immediately after collection. Subsequently, mice were fasted for 24 h and then killed using a cervical dislocation method, after which the colon (from ileocecal valve to anus) was dissected. The morphological changes of colonic mucosa were analyzed using the microscope. The most obvious site of ulceration or erosion was selected, then dissected along the longitudinal axis and fixed with 10% formaldehyde; the rest of the tissue was frozen in liquid nitrogen and stored at − 80 °C.
Immunohistochemistry
The presence of HPSE protein was analyzed using an SP-9000 SPlink detection kit (Zhongshanjinqiao Biotechnology, Beijing, China) following the manufacturer’s instructions. Tissue sections were de-waxed, hydrated and heated. The antigen was retrieved using citric acid buffer; endogenous peroxidase was blocked using goat serum. Samples were then incubated with rabbit polyclonal HPSE antibody (Abcam Inc., Cambridge, MA, USA) at dilution of 1:200 at 4 °C overnight. PBS was used as negative control. Subsequently, samples were incubated with HRP-labeled goat anti-rabbit II antibody (Abcam Inc., Cambridge, MA, USA) at dilution of 1:50 incubation for 1 h. Diaminobenzidine (DAB) was used as a developer, and it was restained with hematoxylin. A light microscope (Nikon eclipse Ci, Japan) was used to analyze the samples.
A double-blind method was used for all sections. Two pathologists read and scored independently. The average value was taken as the final score. Three high power visual fields (× 400 times) were randomly selected for each slice. Semiquantitative integral method was used to make a comprehensive score according to the staining intensity and the proportion of chromogenic cells. ① Tissue color rendering: 0, without color; 1, light yellow or yellow color; 2, brownish yellow color; 3, dark brown color; finally, the average score of three fields of vision was calculated. ② Proportion of staining of target cells (epithelial cells, inflammatory cells and tumor cells): 0, < 5%; 1, 5–25%; 2, 26–50%; 3, 51–75%; 4, > 75%. ③ The final score of each index was obtained by adding the staining intensity and the percentage of chromogenic cells: 0–2, negative (−); 3, weak positive (+), 4–5, positive (++); 6–7, strong positive (+++).
Exfoliated Cells Extraction from Human Feces
The samples were processed as previously described [19, 20]. An approximately 5–10 g human fecal sample was homogenized with a buffer consisting of Hanks solution containing 10% fetal bovine serum (PH7.35) at 200 rpm for 1 min using a Stomacher system (Seward, Thetford, UK). The homogenate was then filtered through a 200-mesh screen to remove large insoluble particles and fibers. Next, the solution was mixed with 80 μL of Dynabeads Epithelial Enrich (Invitrogen, Carlsbad, CA, USA) and incubated for 30 min under a gentle rolling condition at room temperature. The mixture was then placed on the magnetic device and incubated on a shaking platform for 15 min at room temperature. The supernatant was removed, and the colonocytes in the pellet were stored at − 80 °C until RNA extraction.
Exfoliated Cells Extraction from Murine Feces
The samples were processed according to the instructions of CD326 (EpCAM) MicroBeads mouse. (Miltenyi Biotec, Bergisch Gladbach Germany). The murine fecal sample was homogenized with a buffer consisting of PBS (phosphate-buffered saline), BSA (pH 7.2, 0.5% bovine serum albumin) and 2 mM EDTA (by diluting 1:20) at 200 rpm for 1 min using a Stomacher system (Seward, Thetford, UK). The homogenate was filtered through a 30-μm nylon mesh to remove cell clumps which might clog the column, and was followed by the addition of 10 μL of CD326 (EpCAM) MicroBeads mouse (130105958). The sample mixture was then well mixed and incubated for 15 min in a refrigerator (4 °C). The mixture on the magnetic device was incubated on a shaking platform for 10 min at room temperature. The magnetic separation procedure was repeated to increase the purity of colonocytes. The supernatant was then removed, and the colonocytes in the pellet were stored at − 80 °C until RNA extraction.
RTq-PCR
RNA was extracted from colonic mucosal samples, and the exfoliated cells isolated from the fecal samples according to the instructions of RNeasy Mini Kit (Qiagen, Valencia, CA, USA). The cDNA was synthesized by First-Strand cDNA synthesis Kit (Invitrogen, Carlsbad, CA,USA), and the expression level of HPSE mRNA was detected by a quantity nova SYBR Green PCR kit (Qiagen, Valencia, CA, USA). The primer sequences were the following: for heparanase, F-5′-TTCGATCCCAAGAAGGAATCAAC-3′, R-5′-GTAGTGATGCCATGTAACTGAATC-3′; for GAPDH, F-5′-CTGGCGCTGAGTACGTCGTG-3′, R-5′-CAGTCTTCTGGGTGGCAGTG-3′.
The q-PCR was performed following the kit instructions. The amplification volume was 10 μL. The reaction conditions were optimized as follows: denaturation at 95 °C for 2 min, denaturation at 95 °C for 5 s, annealing and elongation at 60 °C for 10 s, 45 cycles. Fluorescence quantitative PCR results analysis was calculated using 2−⊿⊿Ct method: The expression of GAPDH was used to normalize that of the HPSE mRNA and the expression level of HPSE mRNA was expressed as 2−⊿⊿Ct, where ∆Ct = Ct (HPSE mRNA)-Ct (GAPDH), ∆∆Ct = ∆Ct (Sample) − ∆Ct (Normal). The relative expression level was one in normal control group, while it was the value of 2−⊿⊿Ct in the sample.
Statistical Analysis
SPSS 25.0 was used for statistical analysis. The results were expressed as mean ± SD. The results of immunohistochemistry were compared by the Mann–Whitney U method. Multiple groups’ means were compared with a single factor analysis of variance, and the comparison among groups was performed with a t test. Pearson correlation coefficient was used for correlation analysis. P value < 0.05 was considered to be statistically significant.
Results
Expression of HPSE Protein in the UC Patients and the Normal Control Group
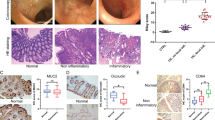
The immunohistochemistry analysis showed no expression or weak expression of HPSE protein in the normal control group (Fig. 1a). In the remission stage of the UC group, the expression of HPSE protein was mainly located in colonic epithelial cells and inflammatory cells, including lymphocytes, neutrophils and plasma cells. The cytoplasm was brownish yellow (Fig. 1b), and its expression was significantly increased in the active stage of the UC group, in both inflammatory cells and in epithelial cells (Fig. 1c). During this stage, the cytoplasm was brownish yellow or dark brown.
Immunohistochemical staining of HPSE protein in the human colonic mucosa and murine colonic mucosa. a Immunohistochemical staining of HPSE protein in the human colonic mucosa in the normal group: The expression of HPSE protein was low in the epithelium of the normal group (magnification × 100). b Immunohistochemical staining of HPSE protein in human colonic mucosa in the remission stage of UC group: The expression of HPSE protein was mainly located in colonic epithelial cells and inflammatory cells, including lymphocytes, neutrophils and histiocytes, and the plasma cells, and the cytoplasm was brownish yellow (magnification × 100). c Immunohistochemical staining of HPSE protein in human colonic mucosa in the active stage of UC group: The expression of HPSE protein was mainly located in colonic epithelial cells and inflammatory cells, including lymphocytes, neutrophils and plasma cells, and the cytoplasm was brownish yellow. The expression of HPSE protein was significantly increased in inflammatory cells and intestinal epithelial cells (magnification × 100). d Immunohistochemical staining of HPSE protein in the murine colonic mucosa in the normal group: The expression of HPSE protein was low in the epithelium of the normal group (magnification × 200). e Immunohistochemical staining of HPSE protein in the murine colonic mucosa in the UC group: The expression of HPSE protein in the UC group was mainly located in inflammatory cells and colonic epithelial cells, and the cytoplasmic staining was brownish yellow (magnification × 200). f Immunohistochemical staining of HPSE protein in murine colonic mucosa in the UCACRC group: The expression of HPSE protein in tumor cells, colonic epithelial cells and inflammatory cells was significantly increased in the UCACRC group (magnification × 200)
The staining results of the normal group were negative or weakly positive. In the UC group, seven cases in the remission stage were positive or weak positive, while 13 cases in the active stage were positive at varying degrees (Table 3). The expression of HPSE protein in the UC group was significantly higher than that in the normal control group (z = 381, P < 0.05, Table 2). Moreover, the expression of HPSE protein in the active stage was significantly higher than that in the remission stage (z = 14, P < 0.05, Table 3).
Relative Expression of HPSE mRNA in Colonic Mucosa and Feces of the UC Patients and the Normal Control Group
In human colonic mucosa, higher expression of HPSE mRNA was detected in the UC group compared to the normal control group (2.690 ± 1.002 vs. 1.018 ± 0.058, P < 0.05). Moreover, the relative expression of HPSE mRNA in the active stage was higher than that in the remission stage (3.151 ± 0.931 vs. 1.833 ± 0.356, P < 0.05, Fig. 2a) and was higher in severe grade of the active stage compared to moderate grade or low grade of the active stage in human colonic mucosa (4.228 ± 0.197 vs. 2.972 ± 0.701 vs. 2.298 ± 0.491, P < 0.05, Fig. 2b). Moreover, the relative expression of HPSE mRNA in human colonic mucosa was significantly correlated with the Mayo score (R2 = 0.633, P < 0.05, Fig. 3a).
Relative expression of HPSE mRNA in human colonic mucosa and human feces of the UC group and the normal control group. a The relative expression of HPSE mRNA in the human colonic mucosa of the UC group and the normal control group. The relative expression of HPSE mRNA in the active stage was significantly higher than that in the remission stage and the normal control group. Mean ± SD, * *P < 0.05 versus the normal control group; #P < 0.05 versus the remission stage. (The normal control group vs. the remission stage, P = 1.090 × 10−5; the normal control group vs. the active stage, P = 7.521 × 10−9; the remission stage vs. the active stage, P = 3.073 × 10−3). b The relative expression of HPSE mRNA in the active stage of the UC group in the human colonic mucosa. The relative expression of HPSE mRNA was higher in severe grade of the active stage compared to moderate grade or low grade of the active stage in human colonic mucosa. Mean ± SD, **P < 0.05 versus the low grade of the active stage, #P < 0.05 versus the moderate grade of the active stage. (The low-grade group vs. the moderate-grade group, P = 1.940 × 10−2; the low-grade group vs. the severe-grade group, P = 7.332 × 10−4; the moderate-grade group vs. the severe-grade group, P = 1.821 × 10−2). c The relative expression of HPSE mRNA in human feces of the UC group and the normal control group. The relative expression of HPSE mRNA of the active stage was significantly higher than that of the remission stage and the normal control groups in human feces. Mean ± SD, **P < 0.05 versus the normal control group; #P < 0.05 versus the remission stage. (The normal control group vs. the remission stage, P = 1.104 × 10−3; the normal control group vs. the active stage, P = 9.023 × 10−6; the remission stage vs. the active stage, P = 2.771 × 10−2). d The relative expression of HPSE mRNA in the active stage of the UC group in human feces. The relative expression of HPSE mRNA was higher in severe grade of the active stage compared to moderate grade or low grade of the active stage in human feces. Mean ± SD, **P < 0.05 versus the low grade of the active stage, #P < 0.05 versus the moderate grade of the active stage. (The low-grade group vs. the moderate-grade group, P = 3.202 × 10−2; the low-grade group vs. the severe-grade group, P = 1.583 × 10−2; the moderate-grade group vs. the severe-grade group, P = 3.552 × 10−2)
Correlation analysis. a The correlation between the relative expression of HPSE mRNA and the Mayo score in human colonic mucosa (R2 = 0.633, P < 0.05). b The correlation between the relative expression of HPSE mRNA and the Mayo score in human feces (R2 = 0.541, P < 0.05). c The correlation between the relative expression of HPSE mRNA in human colonic mucosa and human feces (R2 = 0.634, P < 0.05)
Furthermore, similar data were found in fecal samples. Briefly, the relative expression of HPSE mRNA in the UC group was significantly higher compared to the normal control group in human feces (2.460 ± 0.957 vs. 1.024 ± 0.071, P < 0.05). Moreover, the relative expression of HPSE mRNA in the active stage of the UC group was higher than that in remission stage of the UC group (2.805 ± 1.027 vs. 1.819 ± 0.159, P < 0.05, Fig. 2c), and in severe grade of the active stage compared to moderate grade and low grade of the active stage (3.948 ± 0.962 vs. 2.628 ± 0.249 vs. 1.883 ± 0.475, P < 0.05, Fig. 2d). The relative expression of HPSE mRNA in human feces was significantly correlated with Mayo score (R2 = 0.541, P < 0.05, Fig. 3b). The correlation analysis showed that the content of HPSE mRNA in human feces was significantly correlated with the content of HPSE mRNA in human colonic mucosa (R2 = 0.634, P < 0.05, Fig. 3c).
Expression of HPSE Protein in the Murine Colon with Different Pathological Changes
The immunohistochemistry analysis showed no expression or weak expression of HPSE protein in the normal control group (Fig. 1d). In the UC group, the expression of HPSE protein was mainly located in colonic epithelial cells and inflammatory cells, including lymphocytes, neutrophils and histiocytes, and the cytoplasm was brownish yellow (Fig. 1e). In the UCACRC group, the expression of HPSE protein was significantly increased in tumor cells, inflammatory cells and colonic epithelial cells (Fig. 1f).
Furthermore, the expression of HPSE protein in the UCACRC group was significantly higher than that in the UC group (Z = 316, P < 0.05, Table 4), while the expression of HPSE protein in the UC group was significantly higher than that in the normal control group (Z = 348.5, P < 0.05, Table 5).
Relative Expression of HPSE mRNA in Murine Colonic Mucosa and Murine Feces with Different Pathological Changes
RT-qPCR results showed that the relative expression of HPSE mRNA in murine colonic mucosa of the UCACRC group was 4.229 ± 0.297, which was significantly higher than that of the UC group (3.308 ± 0.187), and the normal control group (1.014 ± 0.067) (P < 0.05, Fig. 4a). The relative expression of HPSE mRNA in murine feces of the UCACRC group was 4.567 ± 0.156, which was significantly higher than that of the UC group (3.577 ± 0.296) and the normal control group (1.065 ± 0.148) (P < 0.05, Fig. 4b). The correlation analysis showed that the relative expression of HPSE mRNA in the murine colonic mucosa of the UCACRC group was significantly correlated with that in the murine feces (R2 = 0.419, P < 0.05, Fig. 4d), and the relative expression of HPSE mRNA in the murine colonic mucosa of the UC group was significantly correlated with that in the murine feces (R2 = 0.362, P < 0.05, Fig. 4c).
Relative expression and correlation analysis of HPSE mRNA in murine colonic mucosa and in murine feces in the normal control group, the UC group, and the UCACRC group. a The relative expression of HPSE mRNA in colonic mucosa of mice in the normal control group, the UC group and the UCACRC group: the relative expression of HPSE mRNA in the UCACRC group was significantly higher than that in the UC group and the normal control group; mean ± SD, * * *P < 0.05 versus the normal control group; #P < 0.05 versus the UC group. (The normal control group vs. the UC group, P = 2.071 × 10−6; the normal control group vs. the UCACRC group, P = 6.220 × 10−7; the UC group vs. the UCACRC group, P = 7.063 × 10−3). b The relative expression of HPSE mRNA in murine feces of the normal control group, the UC group and the UCACRC group: the relative expression of HPSE mRNA in UCACRC group was significantly higher than that in the UC group and the normal control group in feces; mean ± SD, * * *P < 0.05 versus the normal control group; #P < 0.05 versus the UC group. (The normal control group vs. the UC group, P = 1.253 × 10−4; the normal control group vs. the UCACRC group, P = 4.901 × 10−7; the UC group vs. the UCACRC group, P = 1.923 × 10−3). c The correlation between the relative expression of HPSE mRNA in murine colonic mucosa and the relative expression of HPSE mRNA in murine feces of the UC group (R2 = 0.362, P < 0.05). d The correlation between the relative expression of HPSE mRNA in the murine colonic mucosa and the relative expression of HPSE mRNA in murine feces of the UCACRC group (R2 = 0.419, P < 0.05)
Discussion
HPSE may be found in a variety of cells, including macrophages, T and B lymphocytes, endothelial cells, and has a vital role in inflammatory diseases [8]. HPSE proteases can disrupt the structure of the ECM, which, in turn, leads to inflammatory infiltration [13]. HPSE can promote the release of urokinase-type plasminogen activator and tissue-type plasminogen activator, which activates matrix metalloproteinase (MMP) [21]. The degradation of collagen is mainly regulated by different MMPs. The overexpression of MMP has a significant role in the process of tissue damage and repair of the ulcerative colon [22, 23].
HPSE also be found in a wide range of immune cells, which may enhance the function of the adhesion factor. HPSE can help T lymphocytes to adhere to vascular endothelial cells and ECM, which in turn participate in inflammation and regulate the reactivity of immune cells [24]. It is believed that the adaptive immune system composed of T lymphocytes and B lymphocytes has an important role in the occurrence, development and carcinogenesis of ulcerative colitis [25, 26]. Vlodavsky et al. found that HPSE protein and mRNA are highly expressed in the intestinal mucosa of UC patients, but not in the normal intestinal mucosa and the intestinal mucosa of ordinary infectious enteritis [12]. This upregulated expression of HPSE protein and mRNA are closely associated with the non-healing and transformation of chronic colitis to colon cancer [12, 27]. Moreover, Lerner et al. have also confirmed that HPSE and its mRNA continue to be highly expressed throughout ulcerative colitis by establishing animal models. The interaction between HPSE and mucosal macrophages promotes the development of the chronic inflammatory state and the formation of the tumor microenvironment [13]. Yet, the relationship between the expression of HPSE and UC activity has not received adequate attention. In this study, we found that the relative expression of HPSE mRNA was significantly correlated with the Mayo score by comparing the expression of HPSE mRNA in the colonic mucosa of healthy persons and UC patients. Explicitly, the relative expression of HPSE mRNA could be another approach to reflect the activity of ulcerative colitis.
Colorectal disease screening mainly relies on gene detection in fecal samples [7]. It is a noninvasive and does not require any special intestinal preparation and can check all intestinal lesions, which is likely to be a research hot spot in the future [28]. Kanaoka et al. [29] developed a fast and improved detection method based on RT-qPCR that allows the comprehensive analyses of fecal RNA. The detection and analysis of fecal COX-2 mRNA showed that the sensitivity and specificity of COX-2 mRNA expression for the diagnosis of colon cancer were 90% and 100%, respectively. Moreover, Zou H and colleagues [7] found that the expression of hnp1-3 (human neutrophil peptides [HNP]1-3) protein and its mRNA in intestinal mucosa and feces of inflammatory bowel disease and colon cancer increased to varying degrees. The sensitivity of detection of hnp1-3 mRNA expression in feces to inflammatory bowel disease and colon cancer was 80% and 35%, respectively. However, up to now, there are no reports on the detection of HPSE mRNA in feces for prediction of UC activity. Our results revealed a significant correlation between the expression of HPSE mRNA in human feces and human colonic mucosa, suggesting that HPSE mRNA in human feces can reflect the distribution of HPSE mRNA in the human colonic mucosa. Moreover, the expression of HPSE mRNA in human feces was also significantly correlated with the Mayo score, which suggested that HPSE mRNA expression in feces may be used as an indicator of UC activity.
Ulcerative colitis-associated carcinogenesis (UCAC) is a kind of sporadic colon cancer, which is one of the main causes of death in patients with UC. The occurrence of UCAC is closely related to the duration of UC and the extent of lesions. The course of the disease is less than 8–10 years, and the risk of cancer increases by 0.5–1.0% every year, with a high cumulative rate of cancerization (2.5% in 15 years, 13% in 20 years, 34% in 30 years). The patients with a wide range of lesions, such as total colitis, are at a hight risk of developing colon cancer [30]. At present, heparanase has been widely reported to have an essential role in the development and metastasis of colorectal cancer [9,10,11]. However, the role of heparanase in the occurrence and development of UCAC still remains explored. Some scholars have found that the highly coordinated interaction between intestinal heparanase and immune cells (macrophages) can maintain the state of chronic inflammation and create a microenvironment to promote tumor growth and the occurrence and development of UCAC [24]. Furthermore, Lerner et al. confirmed this theory by using heparanase high expression transgenic mice to establish ulcerative colitis carcinogenesis model, which proved that heparanase promotes the formation of tumor microenvironment by activating NF-κ B, STAT3 cell signaling pathways [13]. By setting up a UC-induced carcinogenesis mouse model and comparing the expression of HPSE protein and HPSE mRNA in the normal control group, the UC group and the UCACRC group, we found that the expression level of HPSE protein and HPSE mRNA in the UCACRC group was significantly higher than that in UC group and the normal control group, thus suggesting that HPSE expression has an important role in the carcinogenesis of ulcerative colitis. Moreover, the relative expression levels of HPSE mRNA in murine feces were significantly correlated with that in the murine colonic mucosa. It can be inferred that the detection of HPSE in feces can be used to predict the risk of cancerization of ulcerative colitis.
In conclusion, the detection of HPSE mRNA expression in feces can be used to determine the activity of UC and predict the risk of colon cancer, thus avoiding repeated endoscopy and biopsy. Early detection of HPSE expression in feces is of great significance for population at high risk of UC with a long course of the disease and frequent inflammatory activities. Still the underlying mechanism needs to be further explored.
References
Inflammatory Bowel Disease Group of Digestive Diseases of Chinese Medical Association. The consensus of opinion of inflammatory bowel disease diagnosis and treatment (Beijing, 2018). Chin J Dig. 2018;38:292–311.
Herrinton LJ, Liu L, Levin TR, Allison JE, Lewis JD, Velayos F. Incidence and mortality of colorectal adenocarcinoma in persons with inflammatory bowel disease from 1998 to 2010. Gastroenterology. 2012;143:382–389.
Xiang JY, Qin OY, Guo DL, Xiao NP. Clinical value of fecal calprotectin in determining disease activity of ulcerative colitis. World J Gastroenterol. 2008;14:53–57.
Koga Y, Yasunaga M, Moriya Y, et al. Detection of colorectal cancer cells from faeces using quantitative real-time RT-PCR for colorectal cancer diagnosis. Cancer Sci. 2008;99:1977–1983.
Yang SH, Chien CC, Chen CW, Li SY, Huang CJ. Potential of faecal RNA in diagnosing colorectal cancer. Cancer Lett. 2005;226:55–63.
Ahmed FE, Jeffries CD, Vos PW, et al. Diagnostic microRNA markers for screening sporadic human colon cancer and active ulcerative colitis in stool and tissue. Cancer Genom Proteom. 2009;6:281–295.
Zou H, Harrington JJ, Sugumar A, Klatt KK, Smyrk TC, Ahlquist DA. Detection of colorectal disease by stool defensin assay: an exploratory study. Clin Gastroenterol Hepatol. 2007;5:865–868.
Hao J, Cui M. New advances of heparanase and heparanase-2 in human diseases. Arch Med Res. 2018;49:423–429.
Singh P, Blatt A, Feld S, et al. The heparanase inhibitor PG545 attenuates colon cancer initiation and growth, associating with increased p21 expression. Neoplasia. 2017;19:175–184.
Giordano RJ. Heparanase-2 and syndecan-1 in colon cancer: the ugly ducklings or the beautiful swans? Eur J Gastroenterol Hepatol. 2008;20:716–718.
Kuniyasu H, Chihara Y, Kubozoe T, Takahashi T. Co-expression of CD44v3 and heparanase is correlated with metastasis of human colon cancer. Int J Mol Med. 2002;10:333–337.
Waterman M, Ben-Izhak O, Eliakim R, Groisman G, Vlodavsky I, Ilan N. Heparanase up-regulation by colonic epithelium in inflammatory bowel disease. Mod Pathol. 2007;20:8–14.
Lerner I, Hermano E, Zcharia E, et al. Heparanase powers a chronic inflammatory circuit that promotes colitis-associated tumorigenesis in mice. J Clin Invest. 2011;121:1709–1721.
Lever R, Rose MJ, Mckenzie EA, Page CP. Heparanase induces inflammatory cell recruitment in vivo by promoting adhesion to vascular endothelium. Am J Physiol Cell Physiol. 2014;306:1184–1190.
Wu BW, Li DF, Ke ZF, et al. Expression characteristics of heparanase in colon carcinoma and its close relationship with cyclooxygenase-2 and angiogenesis. Hepatogastroenterology. 2014;104:1510–1514.
Hermano E, Lerner I, Elkin M. Heparanase enzyme in chronic inflammatory bowel disease and colon cancer. Cell Mol Life Sci. 2012;69:2501–2513.
Best WR, Becktel JM, Singleton JW, Kern FJ. Development of Crohn’s disease activity index. National cooperative Crohn’s disease study. Gastroenterology. 1976;70:439–444.
Kohno H, Suzuki R, Sugie S, Tanaka T. Beta-catenin mutations in a mouse model of inflammation-related colon carcinogenesis induced by 1,2-dimethyl hydrazine and dextran sodium sulfate. Cancer Sci. 2005;96:69–76.
Yoshikatsu K, Masahiro Y, Yoshihiro M, et al. Detection of colorectal cancer cells from faeces using quantitative real-time RT-PCR for colorectal cancer diagnosis. Cancer Sci. 2008;9:1977–1983.
Matsushita H, Matsumura Y, Moriya Y, et al. A new method for isolating colonocytes from naturally evacuated feces and its clinical application to colorectal cancer diagnosis. Gastroenterology. 2005;129:1918–1927.
Pirilä E, Ramamurthy NS, Sorsa T, Salo T, Hietanen J, Maisi P. Gelatinase A (MMP-2), collagenase-2 (MMP-8), and laminin-5 gamma2-chain expression in murine inflammatory bowel disease (ulcerative colitis). Dig Dis Sci. 2003;48:93–98. https://doi.org/10.1023/A:1021790532723.
Lin X, Li J, Zhao Q, Feng JR, Gao Q, Nie JY. WGCNA reveals key roles of IL8 and MMP-9 in progression of involvement area in colon of patients with ulcerative colitis. Curr Med Sci. 2018;38:252–258.
Wang YD, Tan XY, Zhang K. Correlation of plasma MMP-1 and TIMP-1 levels and the colonic mucosa expressions in patients with ulcerative colitis. Mediators Inflamm. 2009;2009:275072.
Meirovitz A, Goldberg R, Binder A, Rubinstein AM, Hermano E, Elkin M. Heparanase in inflammation and inflammation- associated cancer. FEBS J. 2013;280:2307–2319.
Shale M, Schiering C, Powrie F. CD4+T cell subsets in intestinal inflammation. Immunol Rev. 2013;252:164–182.
Mayne CG, Williams CB. Induced and natural regulatory T cells in the development of inflammatory bowel disease. Inflamm Bowel Dis. 2013;19:172–178.
Vlodavsky I, Beckhove P, Lerner I, et al. Significance of heparanase in cancer and inflammation. Cancer Microenviron. 2012;5:115–132.
Ahlquist DA, Taylor WR, Mahoney DW, et al. The stool DNA test is more accurate than the plasma septin 9 test in detecting colorectalneoplasia. Clin Gastroenterol Hepatol. 2012;10:272–277.
Kanaoka K, Yoshida KI, Miura N, Sugimura H, Kajimura M. Potential usefulness of detecting cyclooxygenase 2 messenger RNA in feces for colorectal cancer screening. Gastroenterology. 2014;127:422–427.
József S. The pathogenesis of ulcerative colitis-associated colorectal cancer. Magy Onkol. 2004;48:35–43.
Funding
The present study was supported Zhejiang medical and health science and technology project (Number: 2015114978) and Basic public welfare research plan of Zhejiang Province (Number: LGF18H030009).
Author information
Authors and Affiliations
Contributions
Ying Cui performed the experiments and contributed to manuscript preparation. Yan Chen performed experiments and analyzed the data. LC and JZ designed and performed experiments; acquired, analyzed, and interpreted the data; and prepared the manuscript. All authors read and approved the final manuscript. All authors agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval
Our study protocol was approved by the ethical committee of Zhejiang Provincial People’s Hospital (2019KY118). Signed informed consent was obtained from each patient for using materials and publication. The animal studies were approved by the Ethics committee of Zhejiang Chinese Medical University (approval no. IACUC-20180521-03; Hangzhou, China) based on the guidelines for the protection of animals, and all other methods were performed in accordance with the relevant guidelines and regulations.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Cui, Y., Zhang, J., Chen, Y. et al. Upregulation of Fecal Epithelial Heparanase mRNA Is Associated with Increased Ulcerative Colitis Activity and Cancerization Risk. Dig Dis Sci 66, 1488–1498 (2021). https://doi.org/10.1007/s10620-020-06350-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-020-06350-y