Abstract
Background and Aim
Aberrant methylation of specific genes is frequent event in hepatocellular carcinoma (HCC). Our present study aims to explore the methylation levels of acid phosphatase locus 1 (ACP1), bone morphogenetic protein 4 (BMP4), and testis-specific protein, Y-encoded-like 5 (TSPYL5) and their potential clinical applications in HCC.
Methods
The methylation levels of ACP1, BMP4 and TSPYL5 were analyzed in 188 HCC tissues, 163 matched adjacent non-tumor tissues, and 29 normal liver tissues using a method of methylation-sensitive restriction enzyme-based quantitative PCR, and their associations with clinicopathological features and prognosis were evaluated.
Results
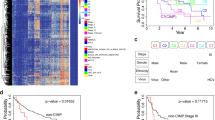
Compared with adjacent non-tumor tissues and normal liver tissues, the methylation levels of ACP1, BMP4, and TSPYL5 were significantly increased in HCC tissues (All p < 0.0001). The methylation of each individual gene could distinguish HCC tissues well from adjacent non-tumor tissues with the area under the receiver operating characteristic curves (AUC) of 0.753, 0.785 and 0.917, respectively. Furthermore, a higher methylation of BMP4 was statistically associated with worse disease-free survival (p = 0.006) and might be an independent unfavorable factor for disease-free survival by univariate and multivariate analysis (p = 0.011, HR 3.431, 95 % CI 1.333–8.833).
Conclusions
Our findings suggest that hypermethylation of ACP1, BMP4, and TSPYL5 are common events in HCC and could be used as potentially detectable biomarkers in HCC tissues. Moreover, BMP4 could be potentially served as a methylated biomarker to predict recurrence and metastasis after hepatectomy for HCC patients. However, their potential clinical application value need to be further clarified.




Similar content being viewed by others
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90.
El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264–1273.e1.
Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255.
Villanueva A, Minguez B, Forner A, Reig M, Llovet JM. Hepatocellular carcinoma: novel molecular approaches for diagnosis, prognosis, and therapy. Annu Rev Med. 2010;61:317–328.
Shibata T, Aburatani H. Exploration of liver cancer genomes. Nat Rev Gastroenterol Hepatol. 2014;11:340–349.
Liu M, Jiang L, Guan XY. The genetic and epigenetic alterations in human hepatocellular carcinoma: a recent update. Protein Cell. 2014;5:673–691.
Um TH, Kim H, Oh BK, et al. Aberrant CpG island hypermethylation in dysplastic nodules and early HCC of hepatitis B virus-related human multistep hepatocarcinogenesis. J Hepatol. 2011;54:939–947.
Nishida N, Nagasaka T, Nishimura T, Ikai I, Boland CR, Goel A. Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology. 2008;47:908–918.
Mah WC, Lee CG. DNA methylation: potential biomarker in hepatocellular carcinoma. Biomark Res. 2014;2:5.
Mikeska T, Bock C, Do H, Dobrovic A. DNA methylation biomarkers in cancer: progress towards clinical implementation. Expert Rev Mol Diagn. 2012;12:473–487.
Villanueva A, Portela A, Sayols S, et al. DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology. 2015;61:1945–1956.
Alho I, Costa L, Bicho M, Coelho C. The role of low-molecular-weight protein tyrosine phosphatase (LMW-PTP ACP1) in oncogenesis. Tumour Biol. 2013;34:1979–1989.
Ricketts CJ, Morris MR, Gentle D, et al. Genome-wide CpG island methylation analysis implicates novel genes in the pathogenesis of renal cell carcinoma. Epigenetics. 2012;7:278–290.
Olk-Batz C, Poetsch AR, Nollke P, et al. Aberrant DNA methylation characterizes juvenile myelomonocytic leukemia with poor outcome. Blood. 2011;117:4871–4880.
Hsu YT, Gu F, Huang YW, et al. Promoter hypomethylation of EpCAM-regulated bone morphogenetic protein gene family in recurrent endometrial cancer. Clin Cancer Res. 2013;19:6272–6285.
Song MA, Tiirikainen M, Kwee S, Okimoto G, Yu H, Wong LL. Elucidating the landscape of aberrant DNA methylation in hepatocellular carcinoma. PLoS One. 2013;8:e55761.
Kim TY, Zhong S, Fields CR, Kim JH, Robertson KD. Epigenomic profiling reveals novel and frequent targets of aberrant DNA methylation-mediated silencing in malignant glioma. Cancer Res. 2006;66:7490–7501.
Jung Y, Park J, Bang YJ, Kim TY. Gene silencing of TSPYL5 mediated by aberrant promoter methylation in gastric cancers. Lab Invest. 2008;88:153–160.
Oka D, Yamashita S, Tomioka T, et al. The presence of aberrant DNA methylation in noncancerous esophageal mucosae in association with smoking history: a target for risk diagnosis and prevention of esophageal cancers. Cancer. 2009;115:3412–3426.
Kim EJ, Lee SY, Kim TR, et al. TSPYL5 is involved in cell growth and the resistance to radiation in A549 cells via the regulation of p21(WAF1/Cip1) and PTEN/AKT pathway. Biochem Biophys Res Commun. 2010;392:448–453.
Shen J, LeFave C, Sirosh I, Siegel AB, Tycko B, Santella RM. Integrative epigenomic and genomic filtering for methylation markers in hepatocellular carcinomas. BMC Med Genomics. 2015;8:28.
Shen J, Wang S, Zhang YJ, et al. Genome-wide DNA methylation profiles in hepatocellular carcinoma. Hepatology. 2012;55:1799–1808.
Ng EK, Leung CP, Shin VY, et al. Quantitative analysis and diagnostic significance of methylated SLC19A3 DNA in the plasma of breast and gastric cancer patients. PLoS One. 2011;6:e22233.
Hua D, Hu Y, Wu YY, et al. Quantitative methylation analysis of multiple genes using methylation-sensitive restriction enzyme-based quantitative PCR for the detection of hepatocellular carcinoma. Exp Mol Pathol. 2011;91:455–460.
Wang S, Dorsey TH, Terunuma A, Kittles RA, Ambs S, Kwabi-Addo B. Relationship between tumor DNA methylation status and patient characteristics in African-American and European-American women with breast cancer. PLoS One. 2012;7:e37928.
Formeister EJ, Tsuchiya M, Fujii H, Shpyleva S, Pogribny IP, Rusyn I. Comparative analysis of promoter methylation and gene expression endpoints between tumorous and non-tumorous tissues from HCV-positive patients with hepatocellular carcinoma. Mutat Res. 2010;692:26–33.
Jain S, Chen S, Chang KC, et al. Impact of the location of CpG methylation within the GSTP1 gene on its specificity as a DNA marker for hepatocellular carcinoma. PLoS One. 2012;7:e35789.
Lou C, Du Z, Yang B, Gao Y, Wang Y, Fang S. Aberrant DNA methylation profile of hepatocellular carcinoma and surgically resected margin. Cancer Sci. 2009;100:996–1004.
Utsunomiya T, Shimada M, Morine Y, Tajima A, Imoto I. Specific molecular signatures of non-tumor liver tissue may predict a risk of hepatocarcinogenesis. Cancer Sci. 2014;105:749–754.
Ramachandran K, Singal R. DNA methylation and field cancerization. Epigenomics. 2012;4:243–245.
Cheng Y, Zhang C, Zhao J, et al. Correlation of CpG island methylator phenotype with poor prognosis in hepatocellular carcinoma. Exp Mol Pathol. 2010;88:112–117.
Tandon P, Garcia-Tsao G. Prognostic indicators in hepatocellular carcinoma: a systematic review of 72 studies. Liver Int. 2009;29:502–510.
Lu X, Zhao H, Yang H, et al. A prospective clinical study on early recurrence of hepatocellular carcinoma after hepatectomy. J Surg Oncol. 2009;100:488–493.
Bruix J, Sherman M, American Association for the Study of Liver D. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020–1022.
Yang T, Lin C, Zhai J, et al. Surgical resection for advanced hepatocellular carcinoma according to Barcelona clinic liver cancer (BCLC) staging. J Cancer Res Clin Oncol. 2012;138:1121–1129.
Acknowledgments
This study was supported by the National Basic Research Program of China (973 Program, 2012CB720605), the Science and Technology Research Plan of Wuhan City (2015060101010057), and Guangdong province production Scientific Research Project (2013B090500101).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None.
Additional information
Xueping Qiu and Bo Hu have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Data, Fig. S1
Representative DNA methylation status of three genes in HCC and paired adjacent non-tumor tissues. (a, b, c, d, e, f) Methylation status of ACP1, BMP4 and TSPYL5 in HCC and paired adjacent non-tumor tissues by BGS. Sequencing for randomly ten clones of PCR products amplified from each sample was shown by black and white circles, which represent methylated and unmethylated CG dinucleotides, respectively. Red circles indicate the CpG sites within recognition sites of MSRE. Each row represents the sequencing result of one clone, and the number represents the position of CpG sites in target regions (number 1 represent the first CpG site in the target region). MSRE–qPCR results for the same sample of BGS were displayed as amplification curve (g, i, k) and melt curve (h, j, l). The blue and purple line indicated digested sample for HCC and paired adjacent non-tumor tissues, respectively. The red and green line indicated undigested sample for HCC and paired adjacent non-tumor tissues, respectively. HCC: hepatocellular carcinoma tissues; NT: adjacent non-tumor tissues (TIFF 1238 kb)
Supplemental Data, Table S1
Primers used in the study. (DOC 35 kb)
Supplemental Data, Table S2
Associations between DNA methylation levels and clinicopathologic parameters. (DOC 91 kb)
Rights and permissions
About this article
Cite this article
Qiu, X., Hu, B., Huang, Y. et al. Hypermethylation of ACP1, BMP4, and TSPYL5 in Hepatocellular Carcinoma and Their Potential Clinical Significance. Dig Dis Sci 61, 149–157 (2016). https://doi.org/10.1007/s10620-015-3878-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-015-3878-3