Abstract
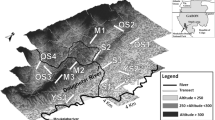
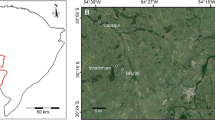
The chamois is a habitat specialist ungulate occupying “continental archipelagos” of fragmented rocky habitats which are frequently restricted to high altitudes. It is not clear whether forest habitats separating such population fragments act as barriers to gene flow. We studied the genetic makeup of the chamois in a topographically diverse landscape at the contact zone of two mountain ranges in Slovenia. Based on sequences of mitochondrial DNA, all Slovenian populations belong to a Northern chamois (Rupicapra r. rupicapra) subspecies. The range of chamois in Slovenia encompasses three different regions, each with unique topography, habitat connectivity and abundance of chamois: the Alps, the Dinaric Mts., and the Pohorje Mts. The habitat of the chamois is extensive and more or less continuous in the Alps, but suboptimal and fragmented in the remaining regions. In agreement with neutral genetic theory, large Northern chamois populations tended to have higher allelic richness and observed heterozygosity. Spatial clustering bears the differentiation into four geographically associated clusters within Slovenia and also revealed a strong substructure within all mountain ranges with suboptimal chamois habitat. Surprisingly, some small Dinaric populations have stayed genetically isolated in restricted habitat patches, even if they are geographically very close to each other. The four clusters, each having a unique demographic history, should be regarded as independent units for management purposes.





Similar content being viewed by others
References
Adamic M, Jerina K (2010) Ungulate management in Europe in the XXI century Slovenia. In: Apollonio M, Andersen R, Putman R (Eds) European ungulates and their management in the 21st century, Cambridge University,Cambride, pp 507–527
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (1996–2004) GENETIX 4.05, logiciel sous Windows TM pour la ge′ne′tique des populations. Laboratoire Ge′nome, Populations, Interactions, CNRS UMR 5000, Universite′ de Montpellier II, Montpellier (France). http://www.genetix.univ-montp2.fr/genetix/intro.htm
Blanco-Aguiar JA, Gonzàlez-Jara P, Ferrero ME, Sànchez-Barbudo I, Virgós E, Villafuerte R, Davila JA (2008) Assessment of game restocking contributions to anthropogenic hybridization: the case of the Iberian red-legged partridge. Anim Conserv 11:535–545
Brook BW, Tonkyn DW, O’Grady JJ, Frankham R (2002) Contribution of inbreeding to extinction risk in threatened species. Ecol Soc 6(1):16–23
Brown JH (2001) Mammals on mountainsides: elevational patterns of diversity. Glob Ecol Biogeogr 10:101–109
Buzan EV, Forster DW, Searle JB, Kryštufek B (2010) A new cytochrome b phylogroup of the common vole (Microtus arvalis) endemic to the Balkans and its implications for the evolutionary history of the species. Biol J Linn Soc 100:788–796
Chapuis M-P, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol 24(3):621–631
Corander J, Sirén J, Arjas E (2008) Bayesian spatial modeling of genetic population structure. Comput Stat 23:111–129
Corlatti L, Lorenzini R, Lovari S (2011) The conservation of the chamois Rupicapra spp. Mammal Rev 41(2):163–174
Crestanello B, Pecchioli E, Vernesi C, Mona C, Martínková N, Janiga M, Hauffe HC, Bertorelle G (2009) The genetic impact of translocations and habitat fragmentation in chamois (Rupicapra spp). J Hered 100:691–708
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Epperson BK (1995) Spatial distributions of genotypes under isolation by distance. Genetics 140:1431–1440
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using Phred I. Accuracy Assess Gen Res 8:175–185
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res 10:564–567
Ezard THG, Travis JMJ (2006) The impact of habitat loss and fragmentation on genetic drift and fixation time. Oikos 114:367–375
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Frantz AC, Tigel Pourtois J, Heuertz M, Schley L, Flamand MC, Krier A, Bertouille S, Chaumont F, Burke T (2006) Genetic structure and assignment tests demonstrate illegal translocation of red deer (Cervus elaphus) into a continuous population. Mol Ecol 15:3191–3203
Frantz AC, Cellina S, Krier A, Schley L, Burke T (2009) Using spatial Bayesian methods to determine the genetic structure of a continuously distributed population: clusters or isolation by distance. J Appl Ecol 46:493–505
Fuchs K, Deutz A, Gressmann G (2000) Detection of space–time clusters and epidemiological examinations of scabies in chamois. Vet Parasitol 92:63–73
Goudet J (2001) FSTAT, a programme to estimate and test gene diversities and fixation indices (version 2.9.3), 2003. http://www2.unil.ch/izea/softwares/fstat.html
Grubb P (2005) Order artiodactyla. In: Wilson DE, Reeder DAM (eds) Mammal species of the world. A taxonomic and geographic reference, vol 1. John Hopkins University, Baltimore, pp 637–743
Guindon S, Gascuel O (2003) A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood”. Sys Biol 52:696–704
Hewitt G (2000) The genetic legacy of the Quaternary ice ages. Nature 405:907–913
Huelsenbeck JP, Ronquist F (2001) MrBayes: Bayesian inference of phylogeny. Bioinformatics 17:754–755
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Kryštufek B (1991) Mammals of Slovenia. Prirodoslovni muzej Slovenije, Ljubljana 294 pp
Kryštufek B (2004) A quantitative assessment of Balkan mammal diversity. In: Griffiths HI, Kryštufek B, Reed JM (eds) Balkan Biodiversity: Process and Pattern in the European hotspot. Kluwer Academic Publ, Dordrecht, NL, pp 79–108
Kryštufek B, Grifiths HI (2003) Anatomy of a human : brown bear conflict. Case study from Slovenia in 1999–2000. In: Kryštfek B, Flajšman B, Griffiths HI (eds) Living with bears: a large European carnivore in a shrinking world. Ljubljana, Ecological form LDS, pp 127–153
Kryštufek B, Milenkovic M, Rapaic Z, Tvrtkovic N (1997) Status and distribution of Caprinae in Former Yugoslavia. In: Shackleton DM (ed) Wild Sheep and Goats and their Relatives. IUCN, Cambridge, UK, pp 138–143
Latch EK, Harveson LA, King JS, Hobson MD, Rhodes OE (2006) Assessing hybridization in wildlife populations using molecular markers: a case study in wild Turkeys. J Wildl Manage 70:485–492
Mares MA, Lacher TE (1987) Ecological, morphological and behavioral convergence in rockdwelling mammals. Curr Mammal 1:307–348
Masini F, Lovari S (1988) Systematics, phylogenetic relationships, and dispersal of the chamois (Rupicapra spp.). Quaternary Res 30:339–349
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Onderscheka K (1982) Etat actuel de la recherche sur la gale du chamois. In: Gindré C (ed) Proceedings: Simposium sur le Chamois. Office National de la Chasse, Ljubljana, pp 90–108
Pérez T, Albornoz J, Domínguez A (2002) Phylogeography of chamois (Rupicapra spp.) inferred from microsatellites. Mol Phylogenet Evol 25:524–534
Rambaud A, Drummond AJ (2007) Tracer, version 1.4. http://beast.bio.ed.ac.uk/Tracer
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Heredity 86:248–249
Rodríguez F, Hammer S, Pérez T, Suchentrunk F, Lorenzini R, Michallet J, Mantinkova N, Albornoz J, Dominguez A (2009) Cytochrome b phylogeography of chamois (Rupicapra spp.). Population contractions, expansions and hybridizations governed the diversification of the genus. J Hered 100:47–55
Rodríguez F, Pérez T, Hammer S, Albornoz J, Domínguez A (2010) Integrating phylogeographic patterns of microsatellite and mtDNA divergence to infer the evolutionary history of chamois. BMC Evol Biol 10:214–222
Ronquist F, Huelsenbeck JP (2003) Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Not 4:137–138
Rossi L, Meneguz PG, DeMartin P, Rodolfi M (1995) The epizootiology of sarcoptic mange in chamois Rupicapra rupicapra, from the Italian Eastern Alps. Parassitologia 37:233–240
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145(4):1219–1228
Rousset F (2008) Genepop’007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Resources 8:103–106
Schaschl H, Kaulfus D, Hammer S, Suchentrunk F (2003) Spatial patterns of mitochondrial and nuclear gene pools in chamois (Rupicapra r. rupicapra) from the Eastern Alps. Heredity 91:125–135
Shackleton DM (1997) Wild sheep and goats and their relatives. International Union for Conservation of Nature, Gland
Soglia D, Rossi L, Cauvin E, Citterio C, Ferroglio E, Maione S, Meneguz PG, Spalenza V, Rasero R, Sacchi P (2010) Population genetic structure of alpine chamois (Rupicapra r. rupicapra) in the Italian Alps. Eur J Wildl Res 56(6):845–854
Spielman D, Brook BW, Frankham R (2004) Most species are not driven to extinction before genetic factors impact them. Proc Natl Acad Sci USA 101:15261–15264
Švigelj L (1961) Medved v Sloveniji (Brown bear in Slovenia). Mladinska knjiga, Ljubljana (in Slovene)
Tamura K, Dudley J, Nei M, Kumar S (2011) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Weir BS, Cockerham CC (1984) Estimating F-Statistics for the analysis of population structure. Evolution 38:1358–1370
Whitlock MC (2003) Fixation probability and time in subdivided populations. Genetics 164:767–779
Willi Y, Van Buskirk J, Schmid B, Fischer M (2006) Genetic isolation of fragmented populations is exacerbated by drift and selection. J Evol Biol 20(2):534–542
Zemanová B, Hájková P, Bryja J, Zima J, Hájková A, Zima J Jr (2011) Development of multiplex microsatellite sets for noninvasive population genetic study of the endangered Tatra chamois. Fol Zool 60:70–80
Acknowledgments
We would like to thank the authorities from the Slovenian Hunting Organisation, Triglav National Park and Rearing Hunting Ground Kozorog, Kamnik for helping to interpret past chamois management. We are grateful to the Hunter Families Log Pod Mangartom, Idrija, Postojna, Ig, Rakitna, Osilnica, Puščava, and Pohorje for collecting the tissue samples. Our thanks also to the Department of Forestry, Biotechnical Faculty, at the University of Ljubljana for providing the map and Peter Glasnović for cartography. Part of the research was sponsored by bilateral project between Slovenia and the Czech Republic (project Contact No: MEB 091023). JB and BZ worked with institutional support RVO: 68081766. We thank Mrs. Karollyn Close for English editing and two anonymous referees for valuable suggestions.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Buzan, E.V., Bryja, J., Zemanová, B. et al. Population genetics of chamois in the contact zone between the Alps and the Dinaric Mountains: uncovering the role of habitat fragmentation and past management. Conserv Genet 14, 401–412 (2013). https://doi.org/10.1007/s10592-013-0469-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-013-0469-8