Abstract
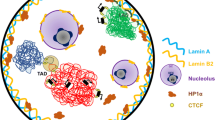
The nucleus is a complex organelle that hosts the genome and is essential for vital processes like DNA replication, DNA repair, transcription, and splicing. The genome is non-randomly organized in the three-dimensional space of the nucleus. This functional sub-compartmentalization was thought to be organized on the framework of nuclear matrix (NuMat), a non-chromatin scaffold that functions as a substratum for various molecular processes of the nucleus. More recently, nuclear bodies or membrane-less subcompartments of the nucleus are thought to arise due to phase separation of chromatin, RNA, and proteins. The nuclear architecture is an amalgamation of the relative organization of chromatin, epigenetic landscape, the nuclear bodies, and the nucleoskeleton in the three-dimensional space of the nucleus. During mitosis, the nucleus undergoes drastic changes in morphology to the degree that it ceases to exist as such; various nuclear components, including the envelope that defines the nucleus, disintegrate, and the chromatin acquires mitosis-specific epigenetic marks and condenses to form chromosome. Upon mitotic exit, chromosomes are decondensed, re-establish hierarchical genome organization, and regain epigenetic and transcriptional status similar to that of the mother cell. How this mitotic memory is inherited during cell division remains a puzzle. NuMat components that are a part of the mitotic chromosome in the form of mitotic chromosome scaffold (MiCS) could potentially be the seeds that guide the relative re-establishment of the epigenome, chromosome territories, and the nuclear bodies. Here, we synthesize the advances towards understanding cellular memory of nuclear architecture across mitosis and propose a hypothesis that a subset of NuMat proteome essential for nucleation of various nuclear bodies are retained in MiCS to serve as seeds of mitotic memory, thus ensuring the daughter cells re-establish the complex status of nuclear architecture similar to that of the mother cells, thereby maintaining the pre-mitotic transcriptional status.



Similar content being viewed by others
Data availability
Not applicable.
Abbreviations
- hnRNP:
-
Heterogenous nuclear ribonucleoprotein
- LAD:
-
Lamina-associated domains
- MiCS:
-
Mitotic chromosome scaffolds
- NuMat:
-
Nuclear matrix
- NOR:
-
Nucleolar-organizing region
- SAF-A:
-
Scaffold attachment factor A
- SAF-B:
-
Scaffold attachment factor B
- S/MARs:
-
Scaffold/matrix-associated regions
- TAD:
-
Topologically associated domains
References
Adolph KW, Cheng SM, Paulson JR, Laemmli UK (1977) Isolation of a protein scaffold from mitotic HeLa cell chromosomes. Proc Natl Acad Sci 74:4937–4941. https://doi.org/10.1073/pnas.74.11.4937
Alabert C, Barth TK, Reverón-Gómez N et al (2015) Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev 29:585–590. https://doi.org/10.1101/gad.256354.114
Alfonso-Parra C, Maggert KA (2010) Drosophila SAF-B links the nuclear matrix, chromosomes, and transcriptional activity. PLOS ONE 5:e10248. https://doi.org/10.1371/journal.pone.0010248
Allen GC, Spiker S, Thompson WF (2000) Use of matrix attachment regions (MARs) to minimize transgene silencing. Plant Mol Biol 43:361–376. https://doi.org/10.1023/A:1006424621037
Amankwaa B, Schoborg T, Labrador M (2022) Drosophila insulator proteins exhibit in vivo liquid–liquid phase separation properties. Life Sci Alliance 5:e202201536. https://doi.org/10.26508/lsa.202201536
Anthony A, Blaxter M (2007) Association of the matrix attachment region recognition signature with coding regions in Caenorhabditis elegans. BMC Genomics 8:418. https://doi.org/10.1186/1471-2164-8-418
Arope S, Harraghy N, Pjanic M, Mermod N (2013) Molecular characterization of a human matrix attachment region epigenetic regulator. PLOS ONE 8:e79262. https://doi.org/10.1371/journal.pone.0079262
Bajpai G, Amiad Pavlov D, Lorber D et al (2021) Mesoscale phase separation of chromatin in the nucleus. eLife 10:e63976. https://doi.org/10.7554/eLife.63976
Banani SF, Lee HO, Hyman AA, Rosen MK (2017) Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18:285–298. https://doi.org/10.1038/nrm.2017.7
Bannister AJ, Kouzarides T (2011) Regulation of chromatin by histone modifications. Cell Res 21:381–395. https://doi.org/10.1038/cr.2011.22
Bártová E, Horáková AH, Uhlířová R et al (2010) Structure and epigenetics of nucleoli in comparison with non-nucleolar compartments. J Histochem Cytochem 58:391–403. https://doi.org/10.1369/jhc.2009.955435
Batty P, Gerlich DW (2019) Mitotic chromosome mechanics: how cells segregate their genome. Trends Cell Biol 29:717–726. https://doi.org/10.1016/j.tcb.2019.05.007
Behera V, Stonestrom AJ, Hamagami N et al (2019) Interrogating histone acetylation and BRD4 as mitotic bookmarks of transcription. Cell Rep 27:400-415.e5. https://doi.org/10.1016/j.celrep.2019.03.057
Bellec M, Dufourt J, Hunt G, et al (2022) The control of transcriptional memory by stable mitotic bookmarking. Nat Commun 13:1176. https://doi.org/10.1038/s41467-022-28855-y
Bellec M, Radulescu O, Lagha M (2018) Remembering the past: mitotic bookmarking in a developing embryo. Curr Opin Syst Biol 11:41–49. https://doi.org/10.1016/j.coisb.2018.08.003
Berezney R, Coffey DS (1974) Identification of a nuclear protein matrix. Biochem Biophys Res Commun 60:1410–1417. https://doi.org/10.1016/0006-291X(74)90355-6
Berezney R, Wei X (1998) The new paradigm: integrating genomic function and nuclear architecture. J Cell Biochem 72:238–242. https://doi.org/10.1002/(SICI)1097-4644(1998)72:30/31+%3c238::AID-JCB29%3e3.0.CO;2-F
Biamonti G, Vourc’h C, (2010) Nuclear stress bodies. Cold Spring Harb Perspect Biol 2:a000695. https://doi.org/10.1101/cshperspect.a000695
Biggs R, Liu PZ, Stephens AD, Marko JF (2019) Effects of altering histone posttranslational modifications on mitotic chromosome structure and mechanics. Mol Biol Cell 30:820–827. https://doi.org/10.1091/mbc.E18-09-0592
Birshtein B (2012) The role of CTCF binding sites in the 3′ immunoglobulin heavy chain regulatory region. Front Genet 3:251. https://doi.org/10.3389/fgene.2012.00251
Bodnar JW, Jones CJ, Coombs DH et al (1983) Proteins tightly bound to HeLa cell DNA at nuclear matrix attachment sites. Mol Cell Biol 3:1567–1579
Boettcher B, Barral Y (2013) The cell biology of open and closed mitosis. Nucleus 4:160–165. https://doi.org/10.4161/nucl.24676
Booth DG, Beckett AJ, Molina O et al (2016) 3D-CLEM reveals that a major portion of mitotic chromosomes is not chromatin. Mol Cell 64:790–802. https://doi.org/10.1016/j.molcel.2016.10.009
Booth DG, Takagi M, Sanchez-Pulido L et al (2014) Ki-67 is a PP1-interacting protein that organises the mitotic chromosome periphery. eLife 3:e01641. https://doi.org/10.7554/eLife.01641
Brahmachari S, Contessoto VG, Di Pierro M, Onuchic JN (2022) Shaping the genome via lengthwise compaction, phase separation, and lamina adhesion. Nucleic Acids Res 50:4258–4271. https://doi.org/10.1093/nar/gkac231
Brugués J, Needleman D (2014) Physical basis of spindle self-organization. Proc Natl Acad Sci 111:18496–18500. https://doi.org/10.1073/pnas.1409404111
Budhavarapu VN, Chavez M, Tyler JK (2013) How is epigenetic information maintained through DNA replication? Epigenetics Chromatin 6:32. https://doi.org/10.1186/1756-8935-6-32
Burgess A, Rasouli M, Rogers S (2014) Stressing mitosis to death. Front. Oncol 4:140
Calikowski TT, Meulia T, Meier I (2003) A proteomic study of the arabidopsis nuclear matrix. J Cell Biochem 90:361–378. https://doi.org/10.1002/jcb.10624
Cavalier-Smith T (1988) Origin of the cell nucleus. BioEssays 9:72–78. https://doi.org/10.1002/bies.950090209
Chen D, Dundr M, Wang C et al (2004) Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J Cell Biol 168:41–54. https://doi.org/10.1083/jcb.200407182
Cheutin T, Cavalli G (2012) Progressive polycomb assembly on H3K27me3 compartments generates polycomb bodies with developmentally regulated motion. PLOS Genet 8:e1002465. https://doi.org/10.1371/journal.pgen.1002465
Chunduri AR, Rajan R, Lima A et al (2022) Dynamics of nuclear matrix attachment regions during 5th instar posterior silk gland development in Bombyx mori. BMC Genomics 23:247. https://doi.org/10.1186/s12864-022-08446-3
Conine CC, Moresco JJ, Gu W et al (2013) Argonautes promote male fertility and provide a paternal memory of germline gene expression in C. elegans. Cell 155:1532–1544. https://doi.org/10.1016/j.cell.2013.11.032
Conte M, Irani E, Chiariello AM et al (2022) Loop-extrusion and polymer phase-separation can co-exist at the single-molecule level to shape chromatin folding. Nat Commun 13:4070. https://doi.org/10.1038/s41467-022-31856-6
Creamer KM, Kolpa HJ, Lawrence JB (2021) Nascent RNA scaffolds contribute to chromosome territory architecture and counter chromatin compaction. Mol Cell 81:3509-3525.e5. https://doi.org/10.1016/j.molcel.2021.07.004
Creamer KM, Lawrence JB (2017) XIST RNA: a window into the broader role of RNA in nuclear chromosome architecture. Philos Trans R Soc B Biol Sci 372:20160360. https://doi.org/10.1098/rstb.2016.0360
Cremer M, von Hase J, Volm T et al (2001) Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res 9:541–567. https://doi.org/10.1023/A:1012495201697
Cuylen S, Blaukopf C, Politi AZ et al (2016) Ki-67 acts as a biological surfactant to disperse mitotic chromosomes. Nature 535:308–312. https://doi.org/10.1038/nature18610
de Belle I, Cai S, Kohwi-Shigematsu T (1998) The genomic sequences bound to special AT-rich sequence-binding protein 1 (SATB1) in vivo in Jurkat T cells are tightly associated with the nuclear matrix at the bases of the chromatin loops. J Cell Biol 141:335–348. https://doi.org/10.1083/jcb.141.2.335
Deluz C, Friman ET, Strebinger D et al (2016) A role for mitotic bookmarking of SOX2 in pluripotency and differentiation. Genes Dev 30:2538–2550. https://doi.org/10.1101/gad.289256.116
Detke S, Keller JM (1982) Comparison of the proteins present in HeLa cell interphase nucleoskeletons and metaphase chromosome scaffolds. J Biol Chem 257:3905–3911. https://doi.org/10.1016/S0021-9258(18)34868-3
Dixon JR, Selvaraj S, Yue F et al (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485:376–380. https://doi.org/10.1038/nature11082
Dobson JR, Hong D, Barutcu AR et al (2017) Identifying nuclear matrix-attached DNA across the genome. J Cell Physiol 232:1295–1305. https://doi.org/10.1002/jcp.25596
Doenecke D (2014) Chromatin dynamics from S-phase to mitosis: contributions of histone modifications. Cell Tissue Res 356:467–475. https://doi.org/10.1007/s00441-014-1873-1
Dorman ER, Bushey AM, Corces VG (2007) The role of insulator elements in large-scale chromatin structure in interphase. Semin Cell Dev Biol 18:682–690. https://doi.org/10.1016/j.semcdb.2007.08.009
Dreval K, Lake RJ, Fan H-Y (2019) HDAC1 negatively regulates selective mitotic chromatin binding of the Notch effector RBPJ in a KDM5A-dependent manner. Nucleic Acids Res 47:4521–4538. https://doi.org/10.1093/nar/gkz178
Dundr M (2012) Nuclear bodies: multifunctional companions of the genome. Curr Opin Cell Biol 24:415–422. https://doi.org/10.1016/j.ceb.2012.03.010
Dundr M, Misteli T (2010) Biogenesis of nuclear bodies. Cold Spring Harb Perspect Biol 2:a000711. https://doi.org/10.1101/cshperspect.a000711
Dundr M, Misteli T (2010) Biogenesis of nuclear bodies. Cold Spring Harb Perspect Biol 2:a000711. https://doi.org/10.1101/cshperspect.a000711
Dykhuizen EC, Hargreaves DC, Miller EL et al (2013) BAF complexes facilitate decatenation of DNA by topoisomerase IIα. Nature 497:624–627. https://doi.org/10.1038/nature12146
Elsing AN, Aspelin C, Björk JK et al (2014) Expression of HSF2 decreases in mitosis to enable stress-inducible transcription and cell survival. J Cell Biol 206:735–749. https://doi.org/10.1083/jcb.201402002
Engelke R, Riede J, Hegermann J et al (2014) The quantitative nuclear matrix proteome as a biochemical snapshot of nuclear organization. J Proteome Res 13:3940–3956. https://doi.org/10.1021/pr500218f
Falk M, Feodorova Y, Naumova N et al (2019) Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature 570:395–399. https://doi.org/10.1038/s41586-019-1275-3
Fan H, Lv P, Huo X et al (2018) The nuclear matrix protein HNRNPU maintains 3D genome architecture globally in mouse hepatocytes. Genome Res 28:192–202. https://doi.org/10.1101/gr.224576.117
Feric M, Misteli T (2021) Phase separation in genome organization across evolution. Trends Cell Biol 31:671–685. https://doi.org/10.1016/j.tcb.2021.03.001
Fey EG, Krochmalnic G, Penman S (1986) The nonchromatin substructures of the nucleus: the ribonucleoprotein (RNP)-containing and RNP-depleted matrices analyzed by sequential fractionation and resinless section electron microscopy. J Cell Biol 102:1654–1665. https://doi.org/10.1083/jcb.102.5.1654
Follmer NE, Wani AH, Francis NJ (2012) A polycomb group protein is retained at specific sites on chromatin in mitosis. PLOS Genet 8:e1003135. https://doi.org/10.1371/journal.pgen.1003135
Fujimura A, Hayashi Y, Kato K et al (2020) Identification of a novel nucleolar protein complex required for mitotic chromosome segregation through centromeric accumulation of Aurora B. Nucleic Acids Res 48:6583–6596. https://doi.org/10.1093/nar/gkaa449
Fujishiro S, Sasai M (2022) Generation of dynamic three-dimensional genome structure through phase separation of chromatin. Proc Natl Acad Sci 119:e2109838119. https://doi.org/10.1073/pnas.2109838119
Galganski L, Urbanek MO, Krzyzosiak WJ (2017) Nuclear speckles: molecular organization, biological function and role in disease. Nucleic Acids Res 45:10350–10368. https://doi.org/10.1093/nar/gkx759
Gallinaro H, Puvion E, Kister L, Jacob M (1983) Nuclear matrix and hnRNP share a common structural constituent associated with premessenger RNA. EMBO J 2:953–960. https://doi.org/10.1002/j.1460-2075.1983.tb01527.x
Ganji M, Shaltiel IA, Bisht S et al (2018) Real-time imaging of DNA loop extrusion by condensin. Science 360:102–105. https://doi.org/10.1126/science.aar7831
Gaszner M, Felsenfeld G (2006) Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet 7:703–713. https://doi.org/10.1038/nrg1925
Gavrilov AA, Zukher IS, Philonenko ES et al (2010) Mapping of the nuclear matrix-bound chromatin hubs by a new M3C experimental procedure. Nucleic Acids Res 38:8051–8060. https://doi.org/10.1093/nar/gkq712
Gerlich D, Beaudouin J, Kalbfuss B et al (2003) Global chromosome positions are transmitted through mitosis in mammalian cells. Cell 112:751–764. https://doi.org/10.1016/S0092-8674(03)00189-2
Getzenberg RH (1994) Nuclear matrix and the regulation of gene expression: tissue specificity. J Cell Biochem 55:22–31. https://doi.org/10.1002/jcb.240550105
Ghotbi E, Ye P, Ervin T et al (2021) Polycomb-group recruitment to a Drosophila target gene is the default state that is inhibited by a transcriptional activator. Sci Adv 7:eabg1556. https://doi.org/10.1126/sciadv.abg1556
Gibney ER, Nolan CM (2010) Epigenetics and gene expression. Heredity 105:4–13. https://doi.org/10.1038/hdy.2010.54
Golbabapour S, Abdulla MA, Hajrezaei M (2011) A concise review on epigenetic regulation: insight into molecular mechanisms. Int J Mol Sci 12:8661–8694. https://doi.org/10.3390/ijms12128661
Goldberg AD, Allis CD, Bernstein E (2007) Epigenetics: a landscape takes shape. Cell 128:635–638. https://doi.org/10.1016/j.cell.2007.02.006
Grasser F, Neusser M, Fiegler H et al (2008) Replication-timing-correlated spatial chromatin arrangements in cancer and in primate interphase nuclei. J Cell Sci 121:1876–1886. https://doi.org/10.1242/jcs.026989
Guelen L, Pagie L, Brasset E et al (2008) Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453:948–951. https://doi.org/10.1038/nature06947
Guillou E, Ibarra A, Coulon V et al (2010) Cohesin organizes chromatin loops at DNA replication factories. Genes Dev 24:2812–2822. https://doi.org/10.1101/gad.608210
Guo J (2014) Transcription: the epicenter of gene expression. J Zhejiang Univ Sci B 15:409–411. https://doi.org/10.1631/jzus.B1400113
Gurudatta B, Yang J, Van Bortle K et al (2013) Dynamic changes in the genomic localization of DNA replication-related element binding factor during the cell cycle. Cell Cycle 12:1605–1615. https://doi.org/10.4161/cc.24742
Haeckel E (1988) Generelle Morphologie der Organismen: Allgemeine Grundzüge der organischen Formen-Wissenschaft, mechanisch begründet durch die von Charles Darwin reformierte Descendenz-Theorie. Band 1: Allgemeine Anatomie. Band 2: Allgemeine Entwicklungsgeschichte. Berlin, New York: De Gruyter. https://doi.org/10.1515/9783110848281
Haeckel E (1904) The wonders of life: a popular study of biological philosophy. Watts & Co., London
Hall LL, Lawrence JB (2016) RNA as a fundamental component of interphase chromosomes: could repeats prove key? Curr Opin Genet Dev 37:137–147. https://doi.org/10.1016/j.gde.2016.04.005
Halsall JA et al. (2021) Histone modifications form a cell-type-specific chromosomal bar code that persists through the cell cycle | Scientific Reports. https://www.nature.com/articles/s41598-021-82539-z
Harr JC, Luperchio TR, Wong X et al (2015) Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. J Cell Biol 208:33–52. https://doi.org/10.1083/jcb.201405110
Hasegawa Y, Brockdorff N, Kawano S et al (2010) The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell 19:469–476. https://doi.org/10.1016/j.devcel.2010.08.006
He DC, Nickerson JA, Penman S (1990) Core filaments of the nuclear matrix. J Cell Biol 110:569–580. https://doi.org/10.1083/jcb.110.3.569
Hearst SM, Gilder AS, Negi SS et al (2009) Cajal-body formation correlates with differential coilin phosphorylation in primary and transformed cell lines. J Cell Sci 122:1872–1881. https://doi.org/10.1242/jcs.044040
Helbig R, Fackelmayer FO (2003) Scaffold attachment factor A (SAF-A) is concentrated in inactive X chromosome territories through its RGG domain. Chromosoma 112:173–182. https://doi.org/10.1007/s00412-003-0258-0
Hernandez-Verdun D (2011) Assembly and disassembly of the nucleolus during the cell cycle. Nucleus 2:189–194. https://doi.org/10.4161/nucl.2.3.16246
Higgins JMG, Prendergast L (2016) Mitotic mysteries: The Case of HP1. Dev Cell 36:477–478. https://doi.org/10.1016/j.devcel.2016.02.019
Hilbert L, Sato Y, Kuznetsova K et al (2021) Transcription organizes euchromatin via microphase separation. Nat Commun 12:1360. https://doi.org/10.1038/s41467-021-21589-3
Hirano T (2012) Condensins: universal organizers of chromosomes with diverse functions. Genes Dev 26:1659–1678. https://doi.org/10.1101/gad.194746.112
Hirota T, Gerlich D, Koch B et al (2004) Distinct functions of condensin I and II in mitotic chromosome assembly. J Cell Sci 117:6435–6445. https://doi.org/10.1242/jcs.01604
Hnilica LS (ed) (2018) Chromosomal nonhistone proteins: structural associations. CRC Press, Boca Raton
Huang Y, Li T, Ems-McClung SC et al (2017) Aurora A activation in mitosis promoted by BuGZ. J Cell Biol 217:107–116. https://doi.org/10.1083/jcb.201706103
Huo X, Ji L, Zhang Y et al (2020) The nuclear matrix protein SAFB cooperates with major satellite RNAs to stabilize heterochromatin architecture partially through phase separation. Mol Cell 77:368-383.e7. https://doi.org/10.1016/j.molcel.2019.10.001
Jenke ACW, Stehle IM, Herrmann F et al (2004) Nuclear scaffold/matrix attached region modules linked to a transcription unit are sufficient for replication and maintenance of a mammalian episome. Proc Natl Acad Sci 101:11322–11327. https://doi.org/10.1073/pnas.0401355101
Jiang H, Wang S, Huang Y et al (2015) Phase transition of spindle-associated protein regulate spindle apparatus assembly. Cell 163:108–122. https://doi.org/10.1016/j.cell.2015.08.010
Kadauke S, Blobel GA (2013) Mitotic bookmarking by transcription factors. Epigenetics Chromatin 6:6. https://doi.org/10.1186/1756-8935-6-6
Kallappagoudar S, Varma P, Pathak RU et al (2010) Nuclear matrix proteome analysis of Drosophila melanogaster*. Mol Cell Proteomics 9:2005–2018. https://doi.org/10.1074/mcp.M110.001362
Kara N, Hossain M, Prasanth SG, Stillman B (2015) Orc1 binding to mitotic chromosomes precedes spatial patterning during G1 phase and assembly of the origin recognition complex in human cells*. J Biol Chem 290:12355–12369. https://doi.org/10.1074/jbc.M114.625012
Kaushal A, Mohana G, Dorier J et al (2021) CTCF loss has limited effects on global genome architecture in Drosophila despite critical regulatory functions. Nat Commun 12:1011. https://doi.org/10.1038/s41467-021-21366-2
Khosraviani N, Ostrowski LA, Mekhail K (2019) Roles for non-coding RNAs in spatial genome organization. Front Cell Dev Biol 7:336
King MR, Petry S (2020) Phase separation of TPX2 enhances and spatially coordinates microtubule nucleation. Nat Commun 11:270. https://doi.org/10.1038/s41467-019-14087-0
Kipp M, Göhring F, Ostendorp T et al (2000) SAF-Box, a conserved protein domain that specifically recognizes scaffold attachment region DNA. Mol Cell Biol 20:7480–7489. https://doi.org/10.1128/MCB.20.20.7480-7489.2000
Klenova EM, Nicolas RH, Paterson HF et al (1993) CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol Cell Biol 13:7612–7624. https://doi.org/10.1128/MCB.13.12.7612
Klymenko T, Papp B, Fischle W et al (2006) A polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev 20:1110–1122. https://doi.org/10.1101/gad.377406
Koch L (2018) Chromosome interaction hubs around nuclear bodies. Nat Rev Genet 19:470–471. https://doi.org/10.1038/s41576-018-0026-x
Kohwi M, Lupton JR, Lai S-L et al (2013) Developmentally regulated subnuclear genome reorganization restricts neural progenitor competence in Drosophila. Cell 152:97–108. https://doi.org/10.1016/j.cell.2012.11.049
Kukalev AS, Lobov IB, Percipalle P, Podgornaya OI (2009) SAF-A/hnRNP-U localization in interphase and metaphase. Cytogenet Genome Res 124:288–297. https://doi.org/10.1159/000218133
Kutay U, Jühlen R, Antonin W (2021) Mitotic disassembly and reassembly of nuclear pore complexes. Trends Cell Biol https://doi.org/10.1016/j.tcb.2021.06.011
L Black K, Petruk S, Fenstermaker TK et al (2016) Chromatin proteins and RNA are associated with DNA during all phases of mitosis. Cell Discov 2:1–14. https://doi.org/10.1038/celldisc.2016.38
Laghmach R, Di Pierro M, Potoyan DA (2021) The interplay of chromatin phase separation and lamina interactions in nuclear organization. Biophys J 120:5005–5017. https://doi.org/10.1016/j.bpj.2021.10.012
Lanctôt C, Cheutin T, Cremer M et al (2007) Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet 8:104–115. https://doi.org/10.1038/nrg2041
Lanzuolo C, Sardo FL, Diamantini A, Orlando V (2011) PcG complexes set the stage for epigenetic inheritance of gene silencing in early S phase before replication. PLOS Genet 7:e1002370. https://doi.org/10.1371/journal.pgen.1002370
Larson AG, Narlikar GJ (2018) The role of phase separation in heterochromatin formation, function, and regulation. Biochem 57:2540–2548. https://doi.org/10.1021/acs.biochem.8b00401
Latonen L (2019) Phase-to-phase with nucleoli – stress responses, protein aggregation and novel roles of RNA. Front Cell Neurosci 13:151
Lee SK, Xue Y, Shen W et al (2018) Topoisomerase 3β interacts with RNAi machinery to promote heterochromatin formation and transcriptional silencing in Drosophila. Nat Commun 9:4946. https://doi.org/10.1038/s41467-018-07101-4
Liang K, Woodfin AR, Slaughter BD et al (2015) Mitotic transcriptional activation: clearance of actively engaged pol II via transcriptional elongation control in mitosis. Mol Cell 60:435–445. https://doi.org/10.1016/j.molcel.2015.09.021
Lieberman-Aiden E, van Berkum NL, Williams L et al (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Sci 326:289–293. https://doi.org/10.1126/science.1181369
Lodhi N, Ji Y, Tulin A (2016) Mitotic bookmarking: maintaining post-mitotic reprogramming of transcription reactivation. Curr Mol Biol Rep 2:10–15. https://doi.org/10.1007/s40610-016-0029-3
Loidl P, Eberharter A (1996) Nuclear matrix and the cell cycle. In: Berezney R, Jeon KW (eds) International Review of Cytology. Academic Press, pp 377–403
Loomis RJ, Naoe Y, Parker JB et al (2009) Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol Cell 33:450–461. https://doi.org/10.1016/j.molcel.2009.02.003
Ludérus MEE, de Graaf A, Mattia E et al (1992) Binding of matrix attachment regions to lamin B1. Cell 70:949–959. https://doi.org/10.1016/0092-8674(92)90245-8
Ma H, Siegel AJ, Berezney R (1999) Association of chromosome territories with the nuclear matrix: disruption of human chromosome territories correlates with the release of a subset of nuclear matrix proteins. J Cell Biol 146:531–542. https://doi.org/10.1083/jcb.146.3.531
Maeshima K, Laemmli UK (2003) A two-step scaffolding model for mitotic chromosome assembly. Dev Cell 4:467–480. https://doi.org/10.1016/S1534-5807(03)00092-3
Mao YS, Zhang B, Spector DL (2011) Biogenesis and function of nuclear bodies. Trends Genet 27:295–306. https://doi.org/10.1016/j.tig.2011.05.006
Marenda M, Lazarova E, Gilbert N (2022) The role of SAF-A/hnRNP U in regulating chromatin structure. Curr Opin Genet Dev 72:38–44. https://doi.org/10.1016/j.gde.2021.10.008
Margueron R, Reinberg D (2010) Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet 11:285–296. https://doi.org/10.1038/nrg2752
Margueron R, Reinberg D (2011) The polycomb complex PRC2 and its mark in life. Nature 469:343–349. https://doi.org/10.1038/nature09784
Martin EW, Thomasen FE, Milkovic NM et al (2021) Interplay of folded domains and the disordered low-complexity domain in mediating hnRNPA1 phase separation. Nucleic Acids Res 49:2931–2945. https://doi.org/10.1093/nar/gkab063
Matharu NK, Ahanger SH (2015) Chromatin insulators and topological domains: adding new dimensions to 3D genome architecture. Genes 6:790–811. https://doi.org/10.3390/genes6030790
Meier I (2005) Subnuclear Trafficking and the Nuclear Matrix. In: Nuclear Import and Export in Plants and Animals. Molecular Biology Intelligence Unit. Springer, Boston. https://doi.org/10.1007/0-387-27747-1_3
Meng Y, Yi X, Li X et al (2016) The non-coding RNA composition of the mitotic chromosome by 5′-tag sequencing. Nucleic Acids Res 44:4934–4946. https://doi.org/10.1093/nar/gkw195
Michelotti EF, Sanford S, Levens D (1997) Marking of active genes on mitotic chromosomes. Nature 388:895–899. https://doi.org/10.1038/42282
Michieletto D, Gilbert N (2019) Role of nuclear RNA in regulating chromatin structure and transcription. Curr Opin Cell Biol 58:120–125. https://doi.org/10.1016/j.ceb.2019.03.007
Mirsky AE, Pollister AW (1942) Nucleoproteins of cell nuclei. Proc Natl Acad Sci 28:344–352. https://doi.org/10.1073/pnas.28.9.344
Mirsky AE, Pollister AW (1946) Chromosin, a desoxyribose nucleoprotein complex of the cell nucleus. J Gen Physiol 30:117–148. https://doi.org/10.1085/jgp.30.2.117
Misteli T (2007) Beyond the sequence: cellular organization of genome function. Cell 128:787–800. https://doi.org/10.1016/j.cell.2007.01.028
Miura H, Hiratani I (2022) Cell cycle dynamics and developmental dynamics of the 3D genome: toward linking the two timescales. Curr Opin Genet Dev 73:101898. https://doi.org/10.1016/j.gde.2021.101898
Monneron A, Blobel G, Palade GE (1972) Fractionation of the nucleus by divalent cations. Isolation of nuclear membranes. J Cell Biol 55:104–125. https://doi.org/10.1083/jcb.55.1.104
Müller J, Verrijzer P (2009) Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr Opin Genet Dev 19:150–158. https://doi.org/10.1016/j.gde.2009.03.001
Narwade N, Patel S, Alam A et al (2019) Mapping of scaffold/matrix attachment regions in human genome: a data mining exercise. Nucleic Acids Res 47:7247–7261. https://doi.org/10.1093/nar/gkz562
Ng RK, Gurdon JB (2008) Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol 10:102–109. https://doi.org/10.1038/ncb1674
Nickerson JA (2022) The ribonucleoprotein network of the nucleus: a historical perspective. Curr Opin Genet Dev 75:101940. https://doi.org/10.1016/j.gde.2022.101940
Nickerson JA, Krockmalnic G, Wan KM, Penman S (1997) The nuclear matrix revealed by eluting chromatin from a cross-linked nucleus. Proc Natl Acad Sci 94:4446–4450. https://doi.org/10.1073/pnas.94.9.4446
Nora EP, Lajoie BR, Schulz EG et al (2012) Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485:381–385. https://doi.org/10.1038/nature11049
Nozawa R-S, Boteva L, Soares DC et al (2017) SAF-A regulates interphase chromosome structure through oligomerization with chromatin-associated RNAs. Cell 169:1214-1227.e18. https://doi.org/10.1016/j.cell.2017.05.029
Nozawa R-S, Gilbert N (2019) RNA: nuclear glue for folding the genome. Trends Cell Biol 29:201–211. https://doi.org/10.1016/j.tcb.2018.12.003
Nuebler J, Fudenberg G, Imakaev M et al (2018) Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc Natl Acad Sci 115:E6697–E6706. https://doi.org/10.1073/pnas.1717730115
Ogg SC, Lamond AI (2002) Cajal bodies and coilin—moving towards function. J Cell Biol 159:17–21. https://doi.org/10.1083/jcb.200206111
Ohta S, Bukowski-Wills J-C, Sanchez-Pulido L et al (2010) The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell 142:810–821. https://doi.org/10.1016/j.cell.2010.07.047
Ohta S, Taniguchi T, Sato N et al (2019) Quantitative proteomics of the mitotic chromosome scaffold reveals the association of BAZ1B with chromosomal axes*. Mol Cell Proteomics 18:169–181. https://doi.org/10.1074/mcp.RA118.000923
Ong JY, Torres JZ (2020) Phase separation in cell division. Mol Cell 80:9–20. https://doi.org/10.1016/j.molcel.2020.08.007
Oomen ME, Dekker J (2017) Epigenetic characteristics of the mitotic chromosome in 1D and 3D. Crit Rev Biochem Mol Biol 52:185–204. https://doi.org/10.1080/10409238.2017.1287160
Ottaviano Y, Gerace L (1985) Phosphorylation of the nuclear lamins during interphase and mitosis. J Biol Chem 260:624–632. https://doi.org/10.1016/S0021-9258(18)89778-2
Palozola KC, Donahue G, Liu H et al (2017) Mitotic transcription and waves of gene reactivation during mitotic exit. Science 358:119–122. https://doi.org/10.1126/science.aal4671
Palozola KC, Lerner J, Zaret KS (2019) A changing paradigm of transcriptional memory propagation through mitosis. Nat Rev Mol Cell Biol 20:55–64. https://doi.org/10.1038/s41580-018-0077-z
Park G, Ryu K, Lee J, Kim G, Park TL, Shim H, ... Cho WK (2022) Topology of gene regulatory compartments relative to the nuclear matrix. https://doi.org/10.1101/2022.05.10.491284
Park J-E, Zhang L, Bang JK et al (2019) Phase separation of Polo-like kinase 4 by autoactivation and clustering drives centriole biogenesis. Nat Commun 10:4959. https://doi.org/10.1038/s41467-019-12619-2
Pathak R, Mamillapalli A, Rangaraj N et al (2013) AAGAG repeat RNA is an essential component of nuclear matrix in Drosophila. RNA Biol 10:564–571. https://doi.org/10.4161/rna.24326
Pathak RU, Srinivasan A, Mishra RK (2014) Genome-wide mapping of matrix attachment regions in Drosophila melanogaster. BMC Genomics 15:1022. https://doi.org/10.1186/1471-2164-15-1022
Paulson JR, Laemmli UK (1977) The structure of histone-depleted metaphase chromosomes. Cell 12:817–828. https://doi.org/10.1016/0092-8674(77)90280-X
Peng A, Weber SC (2019) Evidence for and against liquid-liquid phase separation in the nucleus. Non-Coding RNA 5:50. https://doi.org/10.3390/ncrna5040050
Peric-Hupkes D, Meuleman W, Pagie L et al (2010) Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 38:603–613. https://doi.org/10.1016/j.molcel.2010.03.016
Pflumm MF, Botchan MR (2001) Orc mutants arrest in metaphase with abnormally condensed chromosomes. Development 128:1697–1707. https://doi.org/10.1242/dev.128.9.1697
Phillips-Cremins JE, Sauria MEG, Sanyal A et al (2013) Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153:1281–1295. https://doi.org/10.1016/j.cell.2013.04.053
Pirrotta V, Li H-B (2012) A view of nuclear polycomb bodies. Curr Opin Genet Dev 22:101–109. https://doi.org/10.1016/j.gde.2011.11.004
Poirier MG, Marko JF (2002) Mitotic chromosomes are chromatin networks without a mechanically contiguous protein scaffold. Proc Natl Acad Sci 99:15393–15397. https://doi.org/10.1073/pnas.232442599
Poleshko A, Smith CL, Nguyen SC et al (2019) H3K9me2 orchestrates inheritance of spatial positioning of peripheral heterochromatin through mitosis. eLife 8:e49278. https://doi.org/10.7554/eLife.49278
Pratt AJ, MacRae IJ (2009) The RNA-induced silencing complex: a versatile gene-silencing machine*. J Biol Chem 284:17897–17901. https://doi.org/10.1074/jbc.R900012200
Prescott DM, Bender MA (1962) Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp Cell Res 26:260–268. https://doi.org/10.1016/0014-4827(62)90176-3
Puri D, Swamy CVB, Dhawan J, Mishra RK (2021) Comparative nuclear matrix proteome analysis of skeletal muscle cells in different cellular states. Cell Biol Int 45:580–598. https://doi.org/10.1002/cbin.11499
Raccaud M, Friman ET, Alber AB et al (2019) Mitotic chromosome binding predicts transcription factor properties in interphase. Nat Commun 10:487. https://doi.org/10.1038/s41467-019-08417-5
Rai AK, Chen J-X, Selbach M, Pelkmans L (2018) Kinase-controlled phase transition of membraneless organelles in mitosis. Nature 559:211–216. https://doi.org/10.1038/s41586-018-0279-8
Rao SSP, Huntley MH, Durand NC et al (2014) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159:1665–1680. https://doi.org/10.1016/j.cell.2014.11.021
Ridings-Figueroa R, Stewart ER, Nesterova TB et al (2017) The nuclear matrix protein CIZ1 facilitates localization of Xist RNA to the inactive X-chromosome territory. Genes Dev 31:876–888. https://doi.org/10.1101/gad.295907.117
Romig H, Fackelmayer FO, Renz A et al (1992) Characterization of SAF-A, a novel nuclear DNA binding protein from HeLa cells with high affinity for nuclear matrix/scaffold attachment DNA elements. EMBO J 11:3431–3440. https://doi.org/10.1002/j.1460-2075.1992.tb05422.x
Ryba T, Hiratani I, Sasaki T et al (2011) Replication timing: a fingerprint for cell identity and pluripotency. PLOS Comput Biol 7:e1002225. https://doi.org/10.1371/journal.pcbi.1002225
Schmitz ML, Higgins JMG, Seibert M (2020) Priming chromatin for segregation: functional roles of mitotic histone modifications. Cell Cycle 19:625–641. https://doi.org/10.1080/15384101.2020.1719585
Schneider MWG, Gibson BA, Otsuka S et al (2022) A mitotic chromatin phase transition prevents perforation by microtubules. Nature 609:183–190. https://doi.org/10.1038/s41586-022-05027-y
Schneider R, Grosschedl R (2007) Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev 21:3027–3043. https://doi.org/10.1101/gad.1604607
Schuettengruber B, Bourbon H-M, Di Croce L, Cavalli G (2017) Genome regulation by polycomb and trithorax: 70 years and counting. Cell 171:34–57. https://doi.org/10.1016/j.cell.2017.08.002
Seif E, Kang JJ, Sasseville C et al (2020) Phase separation by the polyhomeotic sterile alpha motif compartmentalizes polycomb group proteins and enhances their activity. Nat Commun 11:5609. https://doi.org/10.1038/s41467-020-19435-z
Sexton T, Yaffe E, Kenigsberg E et al (2012) Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148:458–472. https://doi.org/10.1016/j.cell.2012.01.010
Sharp JA, Perea-Resa C, Wang W, Blower MD (2020) Cell division requires RNA eviction from condensing chromosomes. J Cell Biol 219:e201910148. https://doi.org/10.1083/jcb.201910148
Shen W, Wang D, Ye B et al (2015) A possible role of Drosophila CTCF in mitotic bookmarking and maintaining chromatin domains during the cell cycle. Biol Res 48:27. https://doi.org/10.1186/s40659-015-0019-6
Shoaib M, Nair N, Sørensen CS (2020) Chromatin landscaping at mitotic exit orchestrates genome function. Front Genet 11:103. https://doi.org/10.3389/fgene.2020.00103
Singh PB, Newman AG (2020) On the relations of phase separation and Hi-C maps to epigenetics. R Soc Open Sci 7:191976. https://doi.org/10.1098/rsos.191976
Smetana K, Steele WJ, Busch H (1963) A nuclear ribonucleoprotein network. Exp Cell Res 31:198–201. https://doi.org/10.1016/0014-4827(63)90169-1
Soufi A, Dalton S (2016) Cycling through developmental decisions: how cell cycle dynamics control pluripotency, differentiation and reprogramming. Dev 143:4301–4311. https://doi.org/10.1242/dev.142075
Stauffer DR, Howard TL, Nyun T, Hollenberg SM (2001) CHMP1 is a novel nuclear matrix protein affecting chromatin structure and cell-cycle progression. J Cell Sci 114:2383–2393. https://doi.org/10.1242/jcs.114.13.2383
Steffen PA, Fonseca JP, Gänger C et al (2013) Quantitative in vivo analysis of chromatin binding of polycomb and trithorax group proteins reveals retention of ASH1 on mitotic chromatin. Nucleic Acids Res 41:5235–5250. https://doi.org/10.1093/nar/gkt217
Stenström L et al (2020) Mapping the nucleolar proteome reveals a spatiotemporal organization related to intrinsic protein disorder. Mol Syst Biol 16:e9469. https://doi.org/10.15252/msb.20209469
Strom AR, Emelyanov AV, Mir M et al (2017) Phase separation drives heterochromatin domain formation. Nature 547:241–245. https://doi.org/10.1038/nature22989
Strübbe G, Popp C, Schmidt A et al (2011) Polycomb purification by in vivo biotinylation tagging reveals cohesin and trithorax group proteins as interaction partners. Proc Natl Acad Sci 108:5572–5577. https://doi.org/10.1073/pnas.1007916108
Sureka R, Mishra R (2021) Identification of evolutionarily conserved nuclear matrix proteins and their prokaryotic origins. J Proteome Res 20:518–530. https://doi.org/10.1021/acs.jproteome.0c00550
Sureka R, Wadhwa R, Thakur SS et al (2018) Comparison of nuclear matrix and mitotic chromosome scaffold proteins in Drosophila S2 cells—transmission of hallmarks of nuclear organization through mitosis*. Mol Cell Proteomics 17:1965–1978. https://doi.org/10.1074/mcp.RA118.000591
Sutani T, Sakata T, Nakato R et al (2015) Condensin targets and reduces unwound DNA structures associated with transcription in mitotic chromosome condensation. Nat Commun 6:7815. https://doi.org/10.1038/ncomms8815
Tan S-N, Sim S-P, Khoo ASB (2018) Matrix association region/scaffold attachment region (MAR/SAR) sequence: its vital role in mediating chromosome breakages in nasopharyngeal epithelial cells via oxidative stress-induced apoptosis. BMC Mol Biol 19:15. https://doi.org/10.1186/s12867-018-0116-5
Teves SS, An L, Bhargava-Shah A et al (2018) A stable mode of bookmarking by TBP recruits RNA polymerase II to mitotic chromosomes. eLife 7:e35621. https://doi.org/10.7554/eLife.35621
Timmers HTM, Verrijzer CP (2017) Mitotic chromosomes: not so silent after all. Dev Cell 43:119–121. https://doi.org/10.1016/j.devcel.2017.10.002
Tripathi V, Song DY, Zong X et al (2012) SRSF1 regulates the assembly of pre-mRNA processing factors in nuclear speckles. Mol Biol Cell 23:3694–3706. https://doi.org/10.1091/mbc.e12-03-0206
Valdeolmillos A, Rufas JS, Suja JA et al (2004) Drosophila cohesins DSA1 and Drad21 persist and colocalize along the centromeric heterochromatin during mitosis. Biol Cell 96:457–462. https://doi.org/10.1016/j.biolcel.2004.04.011
Van Bortle K, Nichols MH, Li L et al (2014) Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol 15:R82. https://doi.org/10.1186/gb-2014-15-5-r82
Varma P, Mishra RK (2011) Dynamics of nuclear matrix proteome during embryonic development in Drosophila melanogaster. J Biosci 36:439–459. https://doi.org/10.1007/s12038-011-9081-6
Verheijen R, van Venrooij W, Ramaekers F (1988) The nuclear matrix: structure and composition. J Cell Sci 90:11–36. https://doi.org/10.1242/jcs.90.1.11
Vihervaara A, Sergelius C, Vasara J et al (2013) Transcriptional response to stress in the dynamic chromatin environment of cycling and mitotic cells. Proc Natl Acad Sci 110:E3388–E3397. https://doi.org/10.1073/pnas.1305275110
Vogel MJ, Peric-Hupkes D, van Steensel B (2007) Detection of in vivo protein–DNA interactions using DamID in mammalian cells. Nat Protoc 2:1467–1478. https://doi.org/10.1038/nprot.2007.148
Vogelmann J, Gall AL, Dejardin S et al (2014) Chromatin insulator factors involved in long-range DNA interactions and their role in the folding of the Drosophila genome. PLOS Genet 10:e1004544. https://doi.org/10.1371/journal.pgen.1004544
Volpe TA, Kidner C, Hall IM et al (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833–1837. https://doi.org/10.1126/science.1074973
Wallace HA, Bosco G (2013) Condensins and 3D organization of the interphase nucleus. Curr Genet Med Rep 1:219–229. https://doi.org/10.1007/s40142-013-0024-4
Wang F, Higgins JMG (2013) Histone modifications and mitosis: countermarks, landmarks, and bookmarks. Trends Cell Biol 23:175–184. https://doi.org/10.1016/j.tcb.2012.11.005
Wang J, Yu H, Ma Q et al (2021) Phase separation of OCT4 controls TAD reorganization to promote cell fate transitions. Cell Stem Cell 28:1868-1883.e11. https://doi.org/10.1016/j.stem.2021.04.023
Wilkins BJ, Rall NA, Ostwal Y et al (2014) A cascade of histone modifications induces chromatin condensation in mitosis. Science 343:77–80. https://doi.org/10.1126/science.1244508
Wilson EB (1899) The structure of protoplasm. Sci 10:33–45. https://doi.org/10.1126/science.10.237.33
Wong MM, Byun JS, Sacta M et al (2014) Promoter-bound p300 complexes facilitate post-mitotic transmission of transcriptional memory. PLOS ONE 9:e99989. https://doi.org/10.1371/journal.pone.0099989
Wong X, Hoskins VE, Melendez-Perez AJ et al (2021) Lamin C is required to establish genome organization after mitosis. Genome Biol 22:305. https://doi.org/10.1186/s13059-021-02516-7
Wood AM, Garza-Gongora AG, Kosak ST (2014) A crowdsourced nucleus: understanding nuclear organization in terms of dynamically networked protein function. Biochim Biophys Acta BBA - Gene Regul Mech 1839:178–190. https://doi.org/10.1016/j.bbagrm.2014.01.003
Woodcock CL, Ghosh RP (2010) Chromatin higher-order structure and dynamics. Cold Spring Harb Perspect Biol 2:a000596. https://doi.org/10.1101/cshperspect.a000596
Woodruff JB, Ferreira Gomes B, Widlund PO et al (2017) The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell 169:1066-1077.e10. https://doi.org/10.1016/j.cell.2017.05.028
Yan J, Enge M, Whitington T et al (2013) Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell 154:801–813. https://doi.org/10.1016/j.cell.2013.07.034
Yang J, Sung E, Donlin-Asp PG, Corces VG (2013) A subset of Drosophila Myc sites remain associated with mitotic chromosomes colocalized with insulator proteins. Nat Commun 4:1464. https://doi.org/10.1038/ncomms2469
Zaidi SK, Young DW, Montecino M et al (2011) Bookmarking the genome: maintenance of epigenetic information*. J Biol Chem 286:18355–18361. https://doi.org/10.1074/jbc.R110.197061
Zaidi SK, Young DW, Montecino MA et al (2010) Mitotic bookmarking of genes: a novel dimension to epigenetic control. Nat Rev Genet 11:583–589. https://doi.org/10.1038/nrg2827
Zaret KS (2014) Genome reactivation after the silence in mitosis: recapitulating mechanisms of development? Dev Cell 29:132–134. https://doi.org/10.1016/j.devcel.2014.04.019
Zbarskiĭ IB (1990). Skeletnye struktury kletochnogo iadra v norme i pri patologii [Skeletal structures of the cell nucleus in the normal state and pathology]. Vestn Akad Med Nauk SSSR (2):8–10.
Zhao K, Harel A, Stuurman N et al (1996) Binding of matrix attachment regions to nuclear lamin is mediated by the rod domain and depends on the lamin polymerization state. FEBS Lett 380:161–164. https://doi.org/10.1016/0014-5793(96)00034-8
Acknowledgements
The authors acknowledge the members of the RKM lab and Shivakumara Manu for carefully reading the manuscript and providing critical feedback.
Funding
Research in the RKM lab is supported by grants from the Council for Scientific and Industrial Research (MLP0139), the Government of India, and the JC Bose fellowship (GAP0466). MS is supported by DBT-SRF fellowship.
Author information
Authors and Affiliations
Contributions
MS wrote and edited the manuscript and created illustrations. AB wrote the review. NH wrote the review. RUP wrote the review. RKM conceptualized and edited the review.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Responsible Editor: Helder Maiato
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Soujanya, M., Bihani, A., Hajirnis, N. et al. Nuclear architecture and the structural basis of mitotic memory. Chromosome Res 31, 8 (2023). https://doi.org/10.1007/s10577-023-09714-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10577-023-09714-y