Abstract
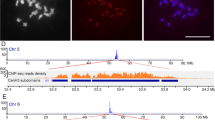
The centromere is a key chromosomal component for sister chromatid cohesion and is the site for kinetochore assembly and spindle fiber attachment, allowing each sister chromatid to faithfully segregate to each daughter cell during cell division. It is not clear what types of sequences act as functional centromeres and how centromere sequences are organized in Oryza brachyantha, an FF genome species. In this study, we found that the three classes of centromere-specific CentO-F satellites (CentO-F1, CentO-F2, and CentO-F3) in O. brachyantha share no homology with the CentO satellites in Oryza sativa. The three classes of CentO-F satellites are all located within the chromosomal regions to which the spindle fibers attach and are characterized by megabase tandem arrays that are flanked by centromere-specific retrotransposons, CRR-F, in the O. brachyantha centromeres. Although these CentO-F satellites are quantitatively variable among 12 O. brachyantha centromeres, immunostaining with an antibody specific to CENH3 indicates that they are colocated with CENH3 in functional centromere regions. Our results demonstrate that the three classes of CentO-F satellites may be the major components of functional centromeres in O. brachyantha.






Similar content being viewed by others
Abbreviations
- BAC:
-
bacterial artificial chromosome
- CentO-F:
-
centromere-specific satellite repeat of FF genome
- CRR-F:
-
centromere-specific retrotransposon of FF genome
- FISH:
-
fluorescence in-situ hybridization
- CEN:
-
centromere of chromosome
References
Aggarwal RK, Brar DS, Khush GS (1997) Two new genomes in the Oryza complex identified on the basis of molecular divergence analysis using total genomic DNA hybridization. Mol Gen Genet 254:1–12
Alfenito MR, Birchler JA (1993) Molecular characterization of a maize B chromosome centric sequence. Genetics 135:589–597
Ammiraju JS, Lu F, Sanyal A, Yu Y, Song X, Jiang N, Pontaroli AC, Rambo T, Currie J, Collura K, Talag J, Fan C, Goicoechea JL, Zuccolo A, Chen J, Bennetzen JL, Chen M, Jackson S, Wing RA (2008) Dynamic evolution of oryza genomes is revealed by comparative genomic analysis of a genus-wide vertical data set. Plant Cell 20:3191–3209
Amor DJ, Kalitsis P, Sumer H, Choo KH (2004) Building the centromere: from foundation proteins to 3D organization. Trends Cell Biol 14:359–368
Bao W, Zhang W, Yang Q, Zhang Y, Han B, Gu M, Xue Y, Cheng Z (2006) Diversity of centromeric repeats in two closely related wild rice species, Oryza officinalis and Oryza rhizomatis. Mol Genet Genom 275:421–430
Bennetzen JL (2007) Patterns in grass genome evolution. Curr Opin Plant Biol 10:176–181
Brandes A, Thompson H, Dean C, Heslop-Harrison JS (1997) Multiple repetitive DNA sequences in the paracentromeric regions of Arabidopsis thaliana L. Chromosom Res 5:238–246
Brar DS, Khush GS (1997) Alien introgression in rice. Plant Mol Biol 35:35–47
Cheng Z, Buell CR, Wing RA, Jiang J (2002a) Resolution of fluorescence in-situ hybridization mapping on rice mitotic prometaphase chromosomes, meiotic pachytene chromosomes and extended DNA fibers. Chromosom Res 10:379–387
Cheng Z, Dong F, Langdon T, Ouyang S, Buell CR, Gu M, Blattner FR, Jiang J (2002b) Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14:1691–1704
Choo KH, Vissel B, Nagy A, Earle E, Kalitsis P (1991) A survey of the genomic distribution of alpha satellite DNA on all the human chromosomes, and derivation of a new consensus sequence. Nucleic Acids Res 19:1179–1182
Clarke L (1990) Centromeres of budding and fission yeasts. Trends Genet 6:150–154
Clarke L (1998) Centromeres: proteins, protein complexes, and repeated domains at centromeres of simple eukaryotes. Curr Opin Genet Dev 8:212–218
Devos KM, Gale MD (1997) Comparative genetics in the grasses. Plant Mol Biol 35:3–15
Dong F, Miller JT, Jackson SA, Wang GL, Ronald PC, Jiang J (1998) Rice (Oryza sativa) centromeric regions consist of complex DNA. Proc Natl Acad Sci U S A 95:8135–8140
Gao D, Gill N, Kim HR, Walling JG, Zhang W, Fan C, Yu Y, Ma J, SanMiguel P, Jiang N, Cheng Z, Wing RA, Jiang J, Jackson SA (2009) A lineage-specific centromere retrotransposon in Oryza brachyantha. Plant J 60:820–831
Ge S, Sang T, Lu BR, Hong DY (1999) Phylogeny of rice genomes with emphasis on origins of allotetraploid species. Proc Natl Acad Sci U S A 96:14400–14405
Hall AE, Keith KC, Hall SE, Copenhaver GP, Preuss D (2004) The rapidly evolving field of plant centromeres. Curr Opin Plant Biol 7:108–114
Hall SE, Kettler G, Preuss D (2003) Centromere satellites from Arabidopsis populations: maintenance of conserved and variable domains. Genome Res 13:195–205
Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF (1997) Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet 15:345–355
Harrison GE, Heslop-Harrison JS (1995) Centromeric repetitive DNA sequences in the genus Brassica. Theor Appl Genet 90:157–165
Hass BL, Pires JC, Porter R, Phillips RL, Jackson SA (2003) Comparative genetics at the gene and chromosome levels between rice ( Oryza sativa) and wildrice ( Zizania palustris). Theor Appl Genet 107:773–782
Henikoff S, Ahmad K, Malik HS (2001) The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293:1098–1102
Jackson SA, Wang ML, Goodman HM, Jiang J (1998) Application of fiber-FISH in physical mapping of Arabidopsis thaliana. Genome 41:566–572
Jiang J, Birchler JA, Parrott WA, Dawe RK (2003) A molecular view of plant centromeres. Trends Plant Sci 8:570–575
Jiang J, Gill BS, Wang GL, Ronald PC, Ward DC (1995) Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc Natl Acad Sci U S A 92:4487–4491
Kang TJ, Yang MS (2004) Rapid and reliable extraction of genomic DNA from various wild-type and transgenic plants. BMC Biotechnol 4:20
Le MH, Duricka D, Karpen GH (1995) Islands of complex DNA are widespread in Drosophila centric heterochromatin. Genetics 141:283–303
Lee HR, Zhang W, Langdon T, Jin W, Yan H, Cheng Z, Jiang J (2005) Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc Natl Acad Sci U S A 102:11793–11798
Lu F, Ammiraju JS, Sanyal A, Zhang S, Song R, Chen J, Li G, Sui Y, Song X, Cheng Z, de Oliveira AC, Bennetzen JL, Jackson SA, Wing RA, Chen M (2009) Comparative sequence analysis of MONOCULM1-orthologous regions in 14 Oryza genomes. Proc Natl Acad Sci U S A 106:2071–2076
Martinez-Zapater JM, Estelle MA, Somerville CR (1986) A highly repeated DNA sequence in Arabidopsis thaliana. Mol Gen Genet 204:417–423
Murata M (2002) Telomeres and Centromeres in Plants. Current Genom 3:527–538
Murphy TD, Karpen GH (1995) Localization of centromere function in a Drosophila minichromosome. Cell 82:599–609
Nagaki K, Cheng Z, Ouyang S, Talbert PB, Kim M, Jones KM, Henikoff S, Buell CR, Jiang J (2004) Sequencing of a rice centromere uncovers active genes. Nat Genet 36:138–145
Nagaki K, Song J, Stupar RM, Parokonny AS, Yuan Q, Ouyang S, Liu J, Hsiao J, Jones KM, Dawe RK, Buell CR, Jiang J (2003) Molecular and cytological analyses of large tracks of centromeric DNA reveal the structure and evolutionary dynamics of maize centromeres. Genetics 163:759–770
Palmer D, Kevany M, Mackworth-Young C, Batchelor R, Lombardi G, Lechler R (1991) Generation and characterization of an HLA-DR alpha-specific monoclonal antibody using L-cell transfectants expressing human and mouse class II major histocompatibility dimers. Immunogenetics 33:12–17
Peterson, D.G., Tomkins, J.P., Frisch, D.A., Wing, R.A., and Paterson, A.H. (2000). Construction of plant bacterial artificial chromosome (BAC) libraries: An illustrated guide. Journal of Agricultural Genomics 5:www.ncgr.org/research/jag
Round EK, Flowers SK, Richards EJ (1997) Arabidopsis thaliana centromere regions: genetic map positions and repetitive DNA structure. Genome Res 7:1045–1053
Singer MF (1982) Highly repeated sequences in mammalian genomes. Int Rev Cytol 76:67–112
Sullivan KF, Hechenberger M, Masri K (1994) Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J Cell Biol 127:581–592
Talbert PB, Bryson TD, Henikoff S (2004) Adaptive evolution of centromere proteins in plants and animals. J Biol 3:18
Talbert PB, Masuelli R, Tyagi AP, Comai L, Henikoff S (2002) Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14:1053–1066
Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH (2008) Synteny and collinearity in plant genomes. Science 320:486–488
Tyler-Smith C, Oakey RJ, Larin Z, Fisher RB, Crocker M, Affara NA, Ferguson-Smith MA, Muenke M, Zuffardi O, Jobling MA (1993) Localization of DNA sequences required for human centromere function through an analysis of rearranged Y chromosomes. Nat Genet 5:368–375
Vafa O, Sullivan KF (1997) Chromatin containing CENP-A and alpha-satellite DNA is a major component of the inner kinetochore plate. Curr Biol 7:897–900
Vaughan DA, Morishima H, Kadowaki K (2003) Diversity in the Oryza genus. Curr Opin Plant Biol 6:139–146
Warburton PE, Cooke CA, Bourassa S, Vafa O, Sullivan BA, Stetten G, Gimelli G, Warburton D, Tyler-Smith C, Sullivan KF, Poirier GG, Earnshaw WC (1997) Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol 7:901–904
Willard HF, Waye JS (1987) Chromosome-specific subsets of human alpha satellite DNA: analysis of sequence divergence within and between chromosomal subsets and evidence for an ancestral pentameric repeat. J Mol Evol 25:207–214
Wolfgruber TK, Sharma A, Schneider KL, Albert PS, Koo DH, Shi J, Gao Z, Han F, Lee H, Xu R, Allison J, Birchler JA, Jiang J, Dawe RK, Presting GG (2009) Maize centromere structure and evolution: sequence analysis of centromeres 2 and 5 reveals dynamic Loci shaped primarily by retrotransposons. PLoS Genet 5:e1000743
Yi CD, Gong ZY, Liang GH, Wang FH, Tang SZ, Gu MH (2007) Isolation and characterization of the centromeric BAC clones from differnet genomes in genus Oryza. HEREDITAS (Beijing) 29:851–858
Zhang D, Yang Q, Bao W, Zhang Y, Han B, Xue Y, Cheng Z (2005a) Molecular cytogenetic characterization of the Antirrhinum majus genome. Genetics 169:325–335
Zhang, W. (2005). Functional Analysis of Centromeric Elements of Two Diploid Genomes BB and FF in Oryza Genus. [D].Beijing:Institute of Genetics and Developmental Biology, Chinese Academy of Sciences
Zhang W, Yi C, Bao W, Liu B, Cui J, Yu H, Cao X, Gu M, Liu M, Cheng Z (2005b) The transcribed 165-bp CentO satellite is the major functional centromeric element in the wild rice species Oryza punctata. Plant Physiol 139:306–315
Zhong CX, Marshall JB, Topp C, Mroczek R, Kato A, Nagaki K, Birchler JA, Jiang J, Dawe RK (2002) Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14:2825–2836
Acknowledgments
This work was supported by grants from the National Basic Research Program of China (“973” Program, 2010CB125904, 2013CBA01405), the National Natural Science Foundation of China (31071382, 30771210) and the Priority Academic Program Development of Jiangsu Higher Education Institutions(PAPD).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Responsible Editor: Hans de Jong.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KF293389-KF293391)
Chuandeng Yi and Wenli Zhang contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 7553 kb)
Rights and permissions
About this article
Cite this article
Yi, C., Zhang, W., Dai, X. et al. Identification and diversity of functional centromere satellites in the wild rice species Oryza brachyantha. Chromosome Res 21, 725–737 (2013). https://doi.org/10.1007/s10577-013-9374-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-013-9374-8