Abstract
Chromosome elimination is a process in which some chromatins are discarded from the presumptive somatic cells during early embryogenesis. Eliminated chromatins in hagfish generally consist of repetitive sequences, and they are highly heterochromatinized in germ cells. In this study, we characterized four novel eliminated DNA families, EEPs1–4, from the Taiwanese hagfish Paramyxine sheni. Sequences of these four elements occupied 20–27% of eliminated DNA in total, and each family was arranged mainly in tandem in the germline genome with high copy numbers. Although most of these elements were eliminated, a minor fraction remained in somatic cells. Some eliminated DNA families are shared as eliminated sequences between Eptatretidae and Myxinidae. Fluorescence in situ hybridization (FISH) of these elements showed that not only heterochromatic chromosomes but also both ends of euchromatic chromosomes in germ cells are absent in somatic cells of P. sheni. It strongly suggests that chromosome terminus elimination, in addition to whole chromosome elimination, contributes to somatic chromosome differentiation. Telomere-FISH further showed that chromosome fragmentation and the subsequent de novo addition of telomeric repeats are the likely mechanisms underlying chromosome terminus elimination. These characteristics make it indispensable to study the evolution and mechanisms underlying chromosome elimination in hagfish.







Similar content being viewed by others
Abbreviations
- Cbs:
-
Chromosome breakage sequence
- CBRs:
-
Chromosomal breakage regions
- DAPI:
-
4′,6-diamidino-2-phenylindole
- DIG:
-
Digoxigenin
- EEEb:
-
Eliminated element of Eptatretus burgeri
- EEEc:
-
Eliminated element of Eptatretus cirrhatus
- EEEo:
-
Eliminated element of Eptatretus okinoseanus
- EEPa:
-
Eliminated element of Paramyxine atami
- EEPs:
-
Eliminated element of Paramyxine sheni
- FISH:
-
Fluorescence in situ hybridization
- FITC:
-
Fluorescein isothiocyanate
- ORF:
-
Open-reading frame
- PI:
-
Propidium iodide
References
Bachmann-Waldmann C, Jentsch S, Tobler H, Müller F (2004) Chromatin diminution leads to rapid evolutionary changes in the organization of the germ line genomes of thenematodes A. suum and P. univalens. Mol Biochem Parasitol 134:53–64
Beermann S (1977) The diminution of Heterochromatic chromosomal segments in Cyclops (Crustacea, Copepoda). Chromosoma 60:297–344
Boveri T (1887) Über Differenzierrung der Zellkerne während der Furchung des Eies von Ascaris megalocephala. Anat Anz 2:688–693
Cammacho JPM, Aharbel TF, Beukeboom LW (2000) B-chromosome evolution. Phil Trans R Soc Lond B 355:163–178
Charlesworth B, Sinegowski P, Stephan W (1994) The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371:215–220
de Saint-Phalle B, Sullivan W (1996) Incomplete sister chromatid separation is the mechanism of programmed chromosome elimination during early Sciara coprophila embryogenesis. Development 122(12):3775–3784
Goto Y, Kubota S, Kohno S (1998) Highly repetitive DNA sequences that are restricted to the germ line in the hagfish Eptatretus cirrhatus: a mosaic of eliminated elements. Chromosoma 107:17–32
Kohno S, Kubota S, Nakai Y (1998) Chromatin diminution and chromosome elimination in hagfish species. In: Jørgensen JM, Lomholt JP, Weber RE, Malte H (eds) The biology of hagfishes. Chapman and Hall, London, pp 81–100
Kubota S, Nakai Y, Kuro-o M, Kohno S (1992) Germ line-restricted supernumerary (B) chromosomes in Eptatretus okinoseanus. Cytogenet Cell Genet 60(3–4):224–228
Kubota S, Kuro-o M, Mizuno S, Kohno S (1993) Germ line-restricted, highly repeated DNA sequences and their chromosomal localization in a Japanese hagfish (Eptatretus okinoseanus). Chromosoma 102(3):163–173
Kubota S, Nakai Y, Sato N, Kuro-o M, Kohno S (1994) Chromosome elimination in northeast Pacific hagfish, Eptatretus stoutii (Cyclostomata, Agnatha). J Heredity 85(5):413–415
Kubota S, Ishibashi T, Kohno S (1997) A germ line-restricted, highly repetitive DNA sequences: an interspecifically conserved, but somatically eliminated, element. Mol Gen Genet 256:252–256
Kubota S, Takano J, Tsuneishi R, Kobayakawa S, Fujikawa N, Nabeyama M, Kohno S (2001) Highly repetitive DNA families restricted to germ cells in a Japanese hagfish (Eptatretus burgeri): a hierarchical and mosaic structure in eliminated chromosomes. Genetica 111:319–328
Kuo CH, Huang S, Lee SC (2003) Phylogeny of hagfish based on the mitochondrial 16S rRNA gene. Mol Phylogenet Evol 28(3):448–457
Kuraku S, Kuratani S (2006) Time scale for cyclostome evolution inferred with a phylogenetic diagnosis of hagfish and lamprey cDNA sequences. Zoolog Sci 23(12):1053–1064
Liu Y, Taverna SD, Muratore TL, Shabanowitz J, Hunt DF, Allis CD (2007) RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev 21(12):1530–1545
Mestrović N, Plohl M, Mravinac B, Ugarković D (1998) Evolution of satellite DNAs from the genus Palorus—experimental evidence for the “library” hypothesis. Mol Biol Evol 15(8):1062–1068
Müller F, Tobler H (2000) Chromatin diminution in the parasitic nematodes Ascaris suum and Parascaris univalens. Int J Parasitol 30(4):391–399
Müller F, Walker P, Aeby P, Neuhaus H, Felder H, Back E, Tobler H (1982) Nucleotide sequence of satellite DNA contained in the eliminated genome of Ascaris lumbricoides. Nucleic Acids Res 10:7493–7510
Nabeyama M, Kubota S, Kohno S (2000) Concerted evolution of a highly repetitive DNA family in Eptatretidae (Cyclostomata, Agnatha) implies specifically differential homogenization and amplification events in their germ cells. J Mol Evol 50:154–169
Nakai Y, Kohno S (1987) Elimination of the largest chromosome pair during differentiation into somatic cells in Japanese hagfish, Myxine garmani (Cyclostomata, Agnatha). Cytogenet Cell Genet 45:80–83
Nakai Y, Kubota S, Kohno S (1991) Chromatin diminution and chromosome elimination in four Japanese hagfish species. Cytogenet Cell Genet 54:196–198
Nakai Y, Kubota S, Goto Y, Ishibashi T, Davison W, Kohno S (1995) Chromosome elimination in three Baltic, south Pacific and northeast Pacific hagfish species. Chromosome Res 3(5):321–330
Niedermaier J, Moritz KB (2000) Organization and dynamics of satellite and telomere DNAs in Ascaris: implications for formation and programmed breakdown of compound chromosomes. Chromosoma 109(7):439–452
Ojima Y (1983) Fish cytogenetics. Suikohsha, Tokyo (in Japanese)
Pons J, Bruvo B, Petitpierre E, Plohl M, Ugarkovic D, Juan C (2004) Complex structural features of satellite DNA sequences in the genus Pimelia (Coleoptera: Tenebrionidae): random differential amplification from a common ‘satellite DNA library’. Heredity 92(5):418–427
Postberg J, Heyse K, Cremer M, Cremer T, Lipps HJ (2008) Spatial and temporal plasticity of chromatin during programmed DNA-reorganization in Stylonychia macronuclear development. Epigenet Chromatin 1(1):3
Salser W, Bowen S, Browne D, el-Adli F, Fedoroff N, Fry K, Heindell H, Paddock G, Poon R, Wallace B, Whitcome P (1976) Investigation of the organization of mammalian chromosomes at the DNA sequence level. Fed Proc 35(1):23–35
Shichiri M, Kikuma Y, Kuo C-H, Liu L-L, Kubota S, Kohno S (1997) Chromosome elimination and germ line-restricted microchromosomes in Paramyxine sheni from Taiwan. Chromosome Sci 1:49–53
Shichiri M, Kubota S, Miyaji K, Kohno S (2001) Ultrastructure of the kinetochores in Japanese hagfish, Eptatretus burgeri. Chromosome Sci 5:139–144
Staiber W (2006) Chromosome elimination in germ line-soma differentiation of Acricotopus lucidus (Diptera, Chironomidae). Genome 49(3):269–274
Streeck RE, Moritz KB, Beer K (1982) Chromatin diminution in Ascaris suum: nucleotide sequence of the eliminated satellite DNA. Nucleic Acids Res 10(11):3495–3502
Taverna SD, Coyne RS, Allis CD (2002) Methylation of histone h3 at lysine 9 targets programmed DNA elimination in tetrahymena. Cell 110(6):701–711
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tobler H (1986) The differentiation of germ and somatic cell lines in nematodes. In: Henning W (ed) Germ line-soma differentiation. Results and problems in cell differentiation 13. Springer, Berlin, pp 1–69
Ugarković D, Plohl M (2002) Variation in satellite DNA profiles—causes and effects. EMBO J 21(22):5955–5959
Yao MC, Chao JL (2005) RNA-guided DNA deletion in Tetrahymena: an RNAi-based mechanism for programmed genome rearrangements. Annu Rev Genet 39:537–559
Yao MC, Duharcourt S, Chalker D (2002) Genome-wide rearrangements of DNA in ciliates. In: Craig N, Craig R, Gellert M, Lambowitz A (eds) Mobile DNA II. ASM, Washington, pp 730–758
Acknowledgments
We are grateful to Mr. Chien-Hsien Kuo and Dr. Li-Lian Liu of the Institute of Marine Biology, National Sun Yat-sen University, Kaohsiung, Taiwan; to Prof. J.-O. Strömberg and the staff of the Kristineberg Marine Biological Station, as well as to Dr. S. Nilsson of the University of Göteborg, Sweden; Dr. W. Davison and Mr. J. van Berkel of the Kaikoura Marine Station, University of Canterbury, New Zealand; Mr. Y. Ono and coworkers at Ono Suisan Ltd., Onahama, Japan; and the staff of the Misaki Marine Biological Station, the University of Tokyo, Japan, for supplying experimental materials. We thank Dr. Mika Fujiwara for useful discussions. This work was supported in part by Nukada Grants-in-Aid for the promotion of scientific research from Toho University, and special grant to S. Kubota in support of advanced scientific projects at the Faculty of Science, Toho University.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Hans J. Lipps
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material S1
Summary of chromosome elimination in hagfishes (DOC 55 kb)
Supplementary material S2
The consensus nucleotide sequence (top) and 34 sequences of EEPs1 family, isolated from the “SmaI fragment”. The consensus sequence was deduced from the 34 repeating units from ten insert sequences in individual clones, designated pUPSS-1, -4, -5, -10, -12, -14, -16, -18, -20, and -21. Dots (.) represent nucleotides identical to those in the consensus sequence at the top. Base substitutions are indicated by the respective bases. Dashes (-) represent alignment gaps. Y indicates C or T. The SmaI and internal HhaI recognition sites are underlined. Box indicates the region in which 16 bp of the palindrome sequence is conserved with 75–87.5% sequence identity. In some unit of each SmaI fragment, palindrome sequences can be extended to almost the complete unit. Arrows denote the inverted repeats. Monomers of EEPs1 were closely related to each other, and the average divergence was 11.6%. The sequence data of pUPSS-12 are available in DDBJ/EMBL/GenBank under accession number AB219575. (JPEG 266 kb)
Supplementary material S3
The consensus nucleotide sequence (top) and 11 sequences of EEPs2 family, isolated from the AluI fragment. The consensus sequence was deduced from the 11 insert sequences in individual clones, designated pUPSA-11, -12, -13, -27, -46, -63, -68, -72, -76, -98, and -99. The AluI and internal Sau 3AI sites were underlined. Arrows denote the direct repeats, each pair sharing more than 77% nucleotide identity. A short sequence motif (positions 71 to 101 in the consensus sequence) showing similarity with the partial sequence of EEPs3 is underlined. Monomers of EEPs2 were closely related to each other, and the average divergence was 2.0%. The sequence data of pUPSA-72 are available in DDBJ/EMBL/GenBank under accession number AB219576. The other notations correspond to those in Supplementary material S2. (JPEG 320 kb)
Supplementary material S4
The consensus nucleotide sequence (top) and 24 sequences of the “Hinf-a” family (EEPs3), isolated from the HinfI fragment. The consensus sequence was deduced from the 19 sequences in individual clones, designated pUPSH-2, -4, -5, -6, -7, -8, -10, -11, -12, -13, -15, -16, -19, -20, -28, -33, -36, -40, and -44. The HinfI recognition sites are underlined, and the three pairs of short direct repeats are denoted by arrows. Each pair of direct repeat shared more than 75% nucleotide identity. Monomers were closely related to one another, and the divergence was 2.5%. The sequence data of pUPSH-13 are available in DDBJ/EMBL/GenBank under accession number AB219577. The other notations correspond to those in Supplementary material S2. (JPEG 188 kb)
Supplementary material S5
The consensus nucleotide sequence (top) and 24 sequences of the “Hinf-b” family (EEPs4), isolated from the HinfI fragment. The consensus sequence was deduced from the 28 repeating units from seven insert sequences in individual clones, designated pUPsH-3, -9, -14, -22, -31, -32, and -38. The average sequence divergence of the 15-bp units was 5.2%. The sequence data of pUPSH-31 are available in DDBJ/EMBL/GenBank under accession number AB219578. The other notations correspond to those in Supplementary material S2. (JPEG 158 kb)
Supplementary material S6
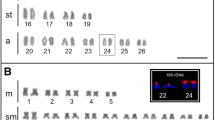
FISH for metaphase chromosomes in primary spermatocytes of P. sheni. FISH signals were detected with DIG-rhodamine (red), while chromatin was counterstained with DAPI (blue). In a and c, only DAPI signals are shown, while in b and d, both DAPI and FISH signals are shown. The FISH probes were pUPSS-14 (EEPs1) for b and pUPSH-13 (EEPs3) for d (Supplementary materials S2 and S4). Asterisks indicate bivalent presumptive somatic chromosomes. Germline-specific chromosomes are smaller than presumptive somatic chromosomes and preferentially show the peripheral distribution. Most of the microchromosomes had accumulated EEPs3 and were strongly stained with DAPI. (JPEG 105 kb)
Supplementary material S7
Localization of the telomeric TTAGGG repeats. Metaphase chromosomes in a spermatogonia, b primary spermatocytes, and c somatic cells of P. sheni after FISH with telomeric repeats [TTAGGG]n as a probe. Hybridization signals of DIG-labeled probes were detected with anti-DIG-fluorescein (yellow). Chromatin was counterstained with PI (red). Bar represents 10 μm. (JPEG 85 kb)
Rights and permissions
About this article
Cite this article
Kojima, N.F., Kojima, K.K., Kobayakawa, S. et al. Whole chromosome elimination and chromosome terminus elimination both contribute to somatic differentiation in Taiwanese hagfish Paramyxine sheni . Chromosome Res 18, 383–400 (2010). https://doi.org/10.1007/s10577-010-9122-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-010-9122-2