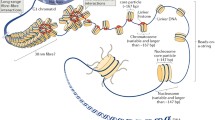
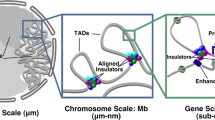
Abstract
Despite a great deal of attention over many years, the structural and functional roles of the linker histone H1 remain enigmatic. The earlier concepts of H1 as a general transcriptional inhibitor have had to be reconsidered in the light of experiments demonstrating a minor effect of H1 deletion in unicellular organisms. More recent work analysing the results of depleting H1 in mammals through genetic knockouts of selected H1 subtypes in the mouse has shown that cells and tissues can tolerate a surprisingly low H1 content. One common feature of H1-depleted nuclei is a reduction in nucleosome repeat length (NRL). Moreover, there is a robust linear relationship between H1 stoichiometry and NRL, suggesting an inherent homeostatic mechanism that maintains intranuclear electrostatic balance. It is also clear that the 1 H1 per nucleosome paradigm for higher eukaryotes is the exception rather than the rule. This, together with the high mobility of H1 within the nucleus, prompts a reappraisal of the role of linker histone as an obligatory chromatin architectural protein.
Similar content being viewed by others
References
Ali T, Thomas JO (2004) Distinct properties of the two putative “globular domains” of the yeast linker histone Hho1p. J Mol Biol 337: 1123–1135.
Allan J, Hartman PG, Crane-Robinson C, Aviles FX (1980) The structure of histone H1 and its location in chromatin. Nature 288: 675–679.
Allan J, Mitchell T, Harborne N, Bohm L, Crane-Robinson (1986) Roles of H1 domains in determining chromatin structure and histone location. J Mol Biol 187: 591–601.
Annunziato AT, Seale RL (1982) Maturation of nucleosomal and non-nucleosomal components of nascent chromatin: differential requirements for concurrent protein synthesis. Biochemistry 21: 5431–5438.
Annunziato AT, Schindler RK, Thomas JA, Seale RL (1981) Dual nature of newly replicated chromatin: evidence for nucleosomal and non-nucleosomal DNA at the site of native replication forks. J Biol Chem 256: 11865–11880.
Ausio J (2000) Are linker histones (histone H1) dispensable for survival? Bioessays 10: 873–877.
Bates DL, Thomas JO (1981) Histones H1 and H5: one or two molecules per nucleosome. Nucl Acids Res 25: 5883–5894.
Bates DL, Butler PJG, Pearson EC, Thomas JO (1981) Stability of the higher order of chicken erythrocyte chromatin in solution. Eur J Biochem 119: 469–476.
Bavykin S, Srebeva L, Banchev T, Tsanev R, Zlatanova J, Mirabekov A (1993) Histone H1 deposition and histone-DNA interactions in replicating chromatin. Proc Natl Acad Sci USA 90: 3918–3922.
Bednar J, Horowitz RA, Grigoryev SA et al. (1998) Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci USA 95: 14173–14178.
Blank TA, Becker PB (1995) Electrostatic mechanism of nucleosome spacing. J Mol Biol 252: 305–313.
Bresnick EH, Bustin M, Marsaud V, Richard-Foy H, Hager GL (1992) The transcriptionally-active MMTV promoter is depleted of histone H1. Nucl Acids Res 20: 5278–5417.
Brown DT (2003) Histone H1 and the dynamic regulation of chromatin function. Biochem Cell Biol 81: 221–227.
Bustin M, Catez F, Lim JH (2005) The dynamics of histone H1 function in chromatin. Mol Cell 17: 617–620.
Caron F, Thomas JO (1981) Exchange of histone H1 between segments of chromatin. J Mol Biol 146: 513–537.
Carruthers LM, Hansen JC (2000) The core histone N-termini function independently of linker histones during chromatin condensation. J Biol Chem 275: 37285–37290.
Carruthers LM, Bednar J, Woodcock CL, Hansen JC (1998) Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays. Biochemistry 37: 14776–14787.
Clark DJ, Kimura T (1990) Electrostatic mechanism of chromatin folding. J Mol Biol 211: 883–896.
Crane-Robinson C (1997) Where is the globular domain of linker histone located on the nucleosome? Trends Biochem Sci 22: 75–77.
Crane-Robinson C (1999) How do linker histones mediate differential gene expression? BioEssays 21: 367–371.
D'Anna JA, Church VL, Tobey RA (1986) Changes in H1 content, nucleosome repeat lengths and DNA elongation under conditions of hydroxyurea treatment that reportedly facilitate gene amplification. Biochim Biophys Acta 868: 137–226.
Dasso M, Dimitrov S, Wolffe AP (1994) Nuclear assembly is independent of linker histones. Proc Natl Acad Sci USA 91: 12477–12481.
Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ (2004) Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science 306: 1571–1573.
Downs JA, Kosmidou E, Morgan A, Jackson SP (2003) Suppression of homologous recombination by the saccharomyces cerevisiae linker histone. Mol Cell 11: 1685–1692.
Fan Y, Sirotkin A, Russell RG, Ayala J, Skoultchi AI (2001) Individual somatic H1 subtypes are dispensable for mouse development even in mice lacking the H1(0) replacement subtype. Mol Cell Biol 21: 7933–7943.
Fan Y, Nikitina T, Morin-Kensicki EM et al. (2003) H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol Cell Biol 23: 4559–4572.
Fan Y, Nikitina T, Zhao J et al. (2005) Depletion of histone H1 in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 123: 1119–1212.
Freidkin I, Katcoff DJ (2001) Specific distribution of the Saccharomyces cerevisiae linker histone homolog Hho1p in the chromatin. Nucl Acids Res 29: 4043–4051.
Georgel P, Hansen JC (2001) Linker histone function in chromatin: dual mechanisms of action. Biochem Cell Biol 79:313–316.
Goytisolo FA, Gerchman SE, Yu X et al. (1996) Identification of two DNA binding sites on the globular domain of histone H5. EMBO J 15: 3421–3429.
Gunjan A, Alexander BA, Sittman DB, Brown DT (1999) Effects of Histone H1 overexpression on chromatin structure. J Biol Chem 53: 37950–37956.
Hamiche A, Schultz P, Ramakrishnan V, Oudet P, Prunell A (1996) Linker histone-dependent DNA structure in linear mononucleosomes. J Mol Biol 257: 30–42.
Harvey AC, Downs JA (2004) What functions do linker histones provide? Mol Microbiol 53: 771–775.
Hellauer K, Sirard E, Turcotte B (2001) Decreased expression of specific genes in yeast cells lacking histone H1. J Biol Chem 276: 13587–13592.
Hendzel MJ, Lever MA, Crawford E, Th'ng, JP (2004) The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J Biol Chem 279: 20028–20034.
Hill DA (2001) Influence of linker histone H1 on chromatin remodeling. Biochem Cell Biol 79: 317–324.
Horn PJ, Carruthers LM, Logie C et al. (2002) Phosphorylation of linker histones regulates ATP-dependent chromatin remodeling enzymes. Nat Struct Biol 9: 263–267.
Kim A, Dean A (2003) A human globin enhancer causes both discrete and widespread alterations in chromatin structure. Mol Cell Biol 23: 8088–8109.
Lever MA, Th'ng JPH, Sun X, Hendzel MJ (2000) Rapid exchange of histone H1.1 on chromatin in living cells. Nature 408: 873–876.
Lu X, Hansen JC (2003) Revisiting the structure and functions of the linker histone C-terminal domain. Biochem Cell Biol 81: 173–176.
Lu X, Hansen JC (2004) Identification of specific functional subdomains within the linker histone H10 C-terminal domain. J Biol Chem 279: 8701–8707.
Manning GS (1978) The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Quart Rev Biophys 11: 179–246.
Maresca TJ, Freedman BS, Heald R (2005) Histone H1 is essential for mitotic chromosome architecture and segregation in Xenopus laevis egg extracts. J Cell Biol 169: 859–869.
Misteli T, Gunjan A, Hock R, Bustin M, Brown DT (2000) Dynamic binding of histone H1 to chromatin in living cells. Nature 408: 877–881.
Morris NR (1976) Nucleosome structure in Aspergillus nidulans. Cell 8: 357–363.
Noll M (1976) Differences and similarities in chromatin structure of Neurospora crassa and higher eukaryotes. Cell 8: 349–355.
Ohsumi K, Katagiri C, Kishimoto T (1993) Chromosome condensation in Xenopus mitotic extracts without H1. Science 262: 2033–2035.
O'Neill TE, Meersseman G, Pennings S, Bradbury EM (1995) Deposition of H1 onto reconstituted nucleosome arrays inhibits both initiation and elongation of transcripts by T7 RNA polymerase. Nucl Acids Res 23: 1075–1082.
Parseghian MH, Hamkalo BA (2001) A compendium of the histone H1 family of somatic subtypes: an elusive cast of characters and their characteristics. Biochem Cell Biol 79: 289–304.
Patterton HG, Landel CC, Landsman D, Petersen CL, Simpson RT (1998) The biochemical and phenotypic characterization of Hho1p, the putative linker histone H1 of Saccharomyces cerevisiae. J Biol Chem 273: 7268–7276.
Pearson EC, Bates DL, Prospero TD, Thomas JO (1984) Neuronal nuclei and glial nuclei from mammalian cerebral cortex. Nucleosome repeat lengths, DNA contents and H1 contents. Eur J Biochem 144: 353–360.
Pennings S, Meerssemann G, Bradbury EM (1994) Linker histones prevent the mobility of positioned nucleosomes. Proc Natl Acad Sci USA 91: 10275–10279.
Pruss D, Bartholomew B, Persinger A et al. (1996) An asymmetric model for the nucleosome: a binding site for linker histones inside the DNA gyres. Science 274: 614–617.
Ramachandran A, Omar M, Cheslock P, Schnitzler GR (2003) Linker histone H1 modulates nucleosome remodeling by human SWI/SNF. J Biol Chem 278: 48590–48601.
Roth SY, Allis CD (1992) Chromatin condensation: does H1 phosphorylation play a role? Trends Biochem Sci 17: 93–98.
Schalch T, Duda S, Sargent DF, Richmond TJ (2005) X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature 436: 138–141.
Shen X, Gorovsky MA (1996) Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell 86: 475–483.
Shen X, Yu L, Weir JW, Gorovsky MA (1995) Linker histones are not essential and affect chromatin condensation in vivo. Cell 82: 47–56.
Shimamura A, Sapp M, Rodriguez-Campos A, Worcel A (1989) Histone H1 represses transcription from minichromosomes assembled in vitro. Mol Cell Biol 9: 5573–5584.
Simpson RT (1978) Structure of the chromatosome, a chromatin core particle containing 160 bp of DNA and all the histones. Biochemistry 17: 5524–5531.
Sirotkin AM, Edelmann W, Cheng G, Klein-Szanto A, Kucherlapati R, Skoultchi AI (1995) Mice develop normally without the H10 linker histone. Proc Natl Acad Sci USA 92: 6434–6438.
Smith CL, Hager GL (1997) Transcrptional regulation of mammalian genes in vivo. J Biol Chem 272: 27493–27496.
Smith PA, Jackson V, Chalkley R (1984) Two-stage maturation process for newly replicated chromatin. Biochemistry 23: 1576–1581.
Stein A, Bina M (1984) A model chromatin assembly system: factors affecting nucleosome spacing. J Mol Biol 178: 341–363.
Thoma F, Koller T, Klug A (1979) Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol 83: 403–427.
Thomas JO (1999) Histone H1: location and role. Curr Opin Cell Biol 11: 312–317.
Thomas JO, Furber V (1976) Yeast chromatin structure. FEBS Lett 66: 274–280.
Travers AA (1999) The location of the linker histone in the nucleosome. Trends Biochem Sci 24: 4–7.
Tremethick DJ, Frommer M (1992) Partial purification, from Xenopus laevis oocytes, of an ATP-dependent activity required for nucleosome spacing in vitro. J Biol Chem 267: 15041–15048.
van Holde KE (1989) Chromatin. New York: Springer-Verlag.
Vignali M, Workman JL (1998) Location and function of linker histones. Natl Struct Biol 5: 1025–1028.
Weintraub H (1985) Histone-H1-dependent chromatin superstructures and the suppression of gene activity. Cell 38: 17–27.
Widom J (1986) Physicochemical studies of the folding of the 100A nucleosomal filament into the 300A filament. Cation dependence. J Mol Biol 190: 411–424.
Widom J (1998) Chromatin structure: linking structure to function with histone H1. Curr Biol 8: R788–R791.
Wolffe AP, Khochbin S, Dimitrov S (1997) What do linker histones do in chromatin? Bioessays 19: 249–255.
Wu M, Allis CD, Richman R, Cook RG, Gorovsky MA (1986) An intervening sequence in an unusual histone H1 gene of Tetrahymena thermophila. Proc Natl Acad Sci USA 83: 8674–8678.
Yuan G-C, Liu Y-J, Dion MF et al. (2005) Genome scale identification of nucleosome positions in S. cerevisiae. Science 309: 626–630.
Zhou YB, Gerchman SE, Ramakrishnan V, Travers A, Muyldermans S (1998) Position and orientation of the globular domain of linker histone H5 on the nucleosome. Nature 395: 402–405.
Zlatanova J, Caiafa P, van Holde K (2000) Linker histone binding and displacement: versatile mechanisms for transcriptional regulation. FASEB J 14: 1697–1704.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Woodcock, C.L., Skoultchi, A.I. & Fan, Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res 14, 17–25 (2006). https://doi.org/10.1007/s10577-005-1024-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-005-1024-3