Abstract
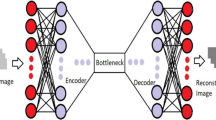
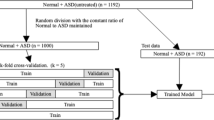
Deep learning (DL) algorithms are increasingly used in cardiac imaging. We aimed to investigate the utility of DL algorithms in de-noising transthoracic echocardiographic images and removing acoustic shadowing artefacts specifically in patients with congenital heart disease (CHD). In addition, the performance of DL algorithms trained on CHD samples was compared to models trained entirely on structurally normal hearts. Deep neural network based autoencoders were built for denoising and removal of acoustic shadowing artefacts based on routine echocardiographic apical 4-chamber views and performance was assessed by visual assessment and quantifying cross entropy. 267 subjects (94 TGA and atrial switch and 39 with ccTGA, 10 Ebstein anomaly, 9 with uncorrected AVSD and 115 normal controls; 56.9% male, age 38.9 ± 15.6 years) with routine transthoracic examinations were included. The autoencoders significantly enhanced image quality across diagnostic subgroups (p < 0.005 for all). Models trained on congenital heart samples performed significantly better when exposed to examples from congenital heart disease patients. Our study demonstrates the potential of autoencoders for denoising and artefact removal in patients with congenital heart disease and structurally normal hearts. While models trained entirely on samples from structurally normal hearts perform reasonably in CHD, our data illustrates the value of dedicated image augmentation systems trained specifically on CHD samples.



Similar content being viewed by others
References
Baumgartner H, Bonhoeffer P, De Groot NM, de Haan F, Deanfield JE, Galie N, Gatzoulis MA, Gohlke-Baerwolf C, Kaemmerer H, Kilner P, Meijboom F, Mulder BJ, Oechslin E, Oliver JM, Serraf A, Szatmari A, Thaulow E, Vouhe PR, Walma E, Task Force on the Management of Grown-up Congenital Heart Disease of the European Society of C, Association for European Paediatric C, Guidelines ESCCfP (2010) ESC guidelines for the management of grown-up congenital heart disease (new version 2010). Eur Heart J 31(23):2915–2957
Li W, West C, McGhie J, van den Bosch AE, Babu-Narayan SV, Meijboom F, Mongeon FP, Khairy P, Kimball TR, Beauchesne LM, Ammash NM, Veldtman GR, Oechslin E, Gatzoulis MA, Webb G (2018) Consensus recommendations for echocardiography in adults with congenital heart defects from the International Society of Adult Congenital Heart Disease (ISACHD). Int J Cardiol 272:77–83
Stout KK, Daniels CJ, Aboulhosn JA, Bozkurt B, Broberg CS, Colman JM, Crumb SR, Dearani JA, Fuller S, Gurvitz M, Khairy P, Landzberg MJ, Saidi A, Valente AM, Van Hare GF (2018) 2018 AHA/ACC guideline for the management of adults with congenital heart disease. Circulation 139:e698–e800
Johnson KW, Torres Soto J, Glicksberg BS, Shameer K, Miotto R, Ali M, Ashley E, Dudley JT (2018) Artificial intelligence in cardiology. J Am Coll Cardiol 71(23):2668–2679
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436–444
Madani A, Arnaout R, Mofrad M, Arnaout R. Fast and accurate view classification of echocardiograms using deep learning. npj Digit Med 2018;1(1):6.
Madani A, Ong JR, Tibrewal A, Mofrad MR. Deep echocardiography: data-efficient supervised and semi-supervised deep learning towards automated diagnosis of cardiac disease. npj Digit Med 2018;1(1):59.
Rajan SP, Kavitha V (2017) Diagnosis of cardiovascular diseases using retinal images through vessel segmentation graph. Curr Med Imaging Rev 13(4):454–459
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, New York, pp 234–241
Salem A-BM, Revett K, El-Dahshan E-SA (2009) Machine learning in electrocardiogram diagnosis. In: International multiconference on computer science and information technology, 2009. IMCSIT'09. IEEE, pp 429–433
Diller GP, Babu-Narayan S, Li W, Radojevic J, Kempny A, Uebing A, Dimopoulos K, Baumgartner H, Gatzoulis M, Orwat S (2019) Utility of machine learning algorithms in assessing patients with a systemic right ventricle. Eur Heart J Cardiovasc Imaging. https://doi.org/10.1093/ehjci/jey211
Bai W, Sinclair M, Tarroni G, Oktay O, Rajchl M, Vaillant G, Lee AM, Aung N, Lukaschuk E, Sanghvi MM, Zemrak F, Fung K, Paiva JM, Carapella V, Kim YJ, Suzuki H, Kainz B, Matthews PM, Petersen SE, Piechnik SK, Neubauer S, Glocker B, Rueckert D (2018) Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. J Cardiovasc Magn Reson 20(1):65
Lang RM, Bierig M, Devereux RB, Flachskampf FA, Foster E, Pellikka PA, Picard MH, Roman MJ, Seward J, Shanewise JS, Solomon SD, Spencer KT, Sutton MS, Stewart WJ, Chamber Quantification Writing G, American Society of Echocardiography's G, Standards C, European Association of E (2005) Recommendations for chamber quantification: a report from the American Society of Echocardiography's Guidelines and Standards Committee and the Chamber Quantification Writing Group, developed in conjunction with the European Association of Echocardiography, a branch of the European Society of Cardiology. J Am Soc Echocardiogr 18(12):1440–1463
Rubinstein R (1999) The cross-entropy method for combinatorial and continuous optimization. Methodol Comput Appl Probab 1(2):127–190
Creswell A, Arulkumaran K, Bharath AA (2017) On denoising autoencoders trained to minimise binary cross-entropy. arXiv:1708.08487
Chollet F (2015) Keras. GitHub.
Turner SP, Monaghan MJ (2006) Tissue harmonic imaging for standard left ventricular measurements: fundamentally flawed? Eur J Echocardiogr 7(1):9–15
Abdi AH, Luong C, Tsang T, Allan G, Nouranian S, Jue J, Hawley D, Fleming S, Gin K, Swift J, Rohling R, Abolmaesumi P (2017) Automatic quality assessment of echocardiograms using convolutional neural networks: feasibility on the apical four-chamber view. IEEE Trans Med Imaging 36(6):1221–1230
Hauptmann A, Arridge S, Lucka F, Muthurangu V, Steeden JA (2018) Real-time cardiovascular MR with spatio-temporal artifact suppression using deep learning—proof of concept in congenital heart disease. Magn Reson Med 81:1143–1156
Acknowledgement
This study was supported by a research Grant from the EMAH Stiftung Karla Voellm, Krefeld, Germany and by the German Competence Network for Congenital Heart Defects (Funded by the German Federal Ministry of Education and Research, BMBF -FKZ 01G10210, 01GI0601).
Funding
This study was supported by a research Grant from the EMAH Stiftung Karla Voellm, Krefeld, Germany. The Adult Congenital Heart Centre and Centre for Pulmonary Hypertension, Royal Brompton Hospital, London, UK have received support from Actelion UK, Pfizer UK, GSK UK, the British Heart Foundation and the NIHR Cardiovascular and Respiratory Biomedical Research Units.
Author information
Authors and Affiliations
Contributions
GPD, AEL and SO planned and conducted the study. GPD, AEL and SO prepared and analyzed the data using DL networks. SBN, RR, WL, MG and HB made substantial contributions in analysis, drafting the article and revising it critically for important intellectual content. All authors gave final approval of the version to be submitted and any revised version.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Diller, GP., Lammers, A.E., Babu-Narayan, S. et al. Denoising and artefact removal for transthoracic echocardiographic imaging in congenital heart disease: utility of diagnosis specific deep learning algorithms. Int J Cardiovasc Imaging 35, 2189–2196 (2019). https://doi.org/10.1007/s10554-019-01671-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10554-019-01671-0