Abstract
Purpose
We conducted an exploratory biomarker study from a phase II clinical trial of eribulin plus gemcitabine (EG) versus paclitaxel plus gemcitabine (PG) in HER2-negative metastatic breast cancer (BC) patients.
Methods
We performed targeted deep sequencing with a customized cancer gene panel and RNA expression assay. Tumor mutation burden (TMB) and mutation signatures were determined based on genetic alteration in targeted regions. Gene set variation analysis was performed with PanCancer Immune Profiling and PanCancer Pathway Panels. Statistical analyses were conducted to identify the associations between genetic alterations and clinical outcomes.
Results
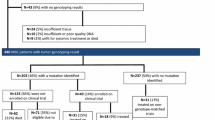
Of 119 patients, 40 had available biomarker data. Among the 40 patients, 4 supported their post-treatment tissues. In targeted deep sequencing, FAT3 (48%) was the most frequently mutated gene, followed by PKHD1, TP53, GATA3, PARP4, and PIK3CA. In terms of gene expression, low expression of epithelial-mesenchymal transition (EMT) pathway genes was associated with prolonged progression-free survival (PFS) in the EG group, while high expression of the EMT pathway was associated with good prognosis in the PG group. Median TMB was 6.5 (range 2.44–46.34) and there was no relationship between TMB and patient prognosis. Analysis of mutation signatures showed that signatures 3, 20, and 26 were frequently observed in our cohort. Further survival analysis according to mutation signature showed that mutation signature 3, as a homologous recombinant deficiency-related signature, was highly associated with disease progression (hazard ratio (log2 scale) 8.21, 95% confidence interval 2.93–13.48, p = 0.002). Kaplan–Meier plot also showed that BCs with signature 3 had short PFS compared to those without these signatures (median PFS (months) for signature 3 (low vs. high): 17.2 vs. 8.1, p = 0.0026).
Conclusions
Mutation signature 3, found in about 30% of MBCs regardless of hormone receptor status, was associated with short PFS for patients with cytotoxic chemotherapy.
Trial registry
ClinicalTrials.gov number: NCT02263495.





Similar content being viewed by others
References
Andre F, Hurvitz S, Fasolo A, Tseng LM, Jerusalem G, Wilks S, O’Regan R, Isaacs C, Toi M, Burris H, He W, Robinson D, Riester M, Taran T, Chen D, Slamon D (2016) Molecular alterations and everolimus efficacy in human epidermal growth factor receptor 2-overexpressing metastatic breast cancers: combined exploratory biomarker analysis From BOLERO-1 and BOLERO-3. J Clin Oncol 34:2115–2124. https://doi.org/10.1200/JCO.2015.63.9161
Cristofanilli M, DeMichele A, Giorgetti C, Turner NC, Slamon DJ, Im SA, Masuda N, Verma S, Loi S, Colleoni M, Theall KP, Huang X, Liu Y, Bartlett CH (2018) Predictors of prolonged benefit from palbociclib plus fulvestrant in women with endocrine-resistant hormone receptor-positive/human epidermal growth factor receptor 2-negative metastatic breast cancer in PALOMA-3. Eur J Cancer 104:21–31. https://doi.org/10.1016/j.ejca.2018.08.011
Hyman DM, Piha-Paul SA, Won H, Rodon J, Saura C, Shapiro GI, Juric D, Quinn DI, Moreno V, Doger B, Mayer IA, Boni V, Calvo E, Loi S, Lockhart AC, Erinjeri JP, Scaltriti M, Ulaner GA, Patel J, Tang J, Beer H, Selcuklu SD, Hanrahan AJ, Bouvier N, Melcer M, Murali R, Schram AM, Smyth LM, Jhaveri K, Li BT, Drilon A, Harding JJ, Iyer G, Taylor BS, Berger MF, Cutler RE Jr, Xu F, Butturini A, Eli LD, Mann G, Farrell C, Lalani AS, Bryce RP, Arteaga CL, Meric-Bernstam F, Baselga J, Solit DB (2018) HER kinase inhibition in patients with HER2- and HER3-mutant cancers. Nature 554:189–194. https://doi.org/10.1038/nature25475
Tutt A, Tovey H, Cheang MCU, Kernaghan S, Kilburn L, Gazinska P, Owen J, Abraham J, Barrett S, Barrett-Lee P, Brown R, Chan S, Dowsett M, Flanagan JM, Fox L, Grigoriadis A, Gutin A, Harper-Wynne C, Hatton MQ, Hoadley KA, Parikh J, Parker P, Perou CM, Roylance R, Shah V, Shaw A, Smith IE, Timms KM, Wardley AM, Wilson G, Gillett C, Lanchbury JS, Ashworth A, Rahman N, Harries M, Ellis P, Pinder SE, Bliss JM (2018) Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: the TNT Trial. Nat Med 24:628–637. https://doi.org/10.1038/s41591-018-0009-7
Litton JK, Rugo HS, Ettl J, Hurvitz SA, Goncalves A, Lee KH, Fehrenbacher L, Yerushalmi R, Mina LA, Martin M, Roche H, Im YH, Quek RGW, Markova D, Tudor IC, Hannah AL, Eiermann W, Blum JL (2018) Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. N Engl J Med 379:753–763. https://doi.org/10.1056/NEJMoa1802905
Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N, Delaloge S, Li W, Tung N, Armstrong A, Wu W, Goessl C, Runswick S, Conte P (2017) Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med 377:523–533. https://doi.org/10.1056/NEJMoa1706450
Pichon MF, Broet P, Magdelenat H, Delarue JC, Spyratos F, Basuyau JP, Saez S, Rallet A, Courriere P, Millon R, Asselain B (1996) Prognostic value of steroid receptors after long-term follow-up of 2257 operable breast cancers. Br J Cancer 73:1545–1551
Kim HK, Lee SH, Kim YJ, Park SE, Lee HS, Lim SW, Cho JH, Kim JY, Ahn JS, Im YH, Yu JH, Park YH (2019) Does guideline non-adherence result in worse clinical outcomes for hormone receptor-positive and HER2-negative metastatic breast cancer in premenopausal women?: result of an institution database from South Korea. BMC Cancer 19:84. https://doi.org/10.1186/s12885-018-5258-9
Bartlett JM, Munro A, Cameron DA, Thomas J, Prescott R, Twelves CJ (2008) Type 1 receptor tyrosine kinase profiles identify patients with enhanced benefit from anthracyclines in the BR9601 adjuvant breast cancer chemotherapy trial. J Clin Oncol 26:5027–5035. https://doi.org/10.1200/JCO.2007.14.6597
Okuma HS, Koizumi F, Hirakawa A, Nakatochi M, Komori O, Hashimoto J, Kodaira M, Yunokawa M, Yamamoto H, Yonemori K, Shimizu C, Fujiwara Y, Tamura K (2016) Clinical and microarray analysis of breast cancers of all subtypes from two prospective preoperative chemotherapy studies. Br J Cancer 115:411–419. https://doi.org/10.1038/bjc.2016.184
Kim JY, Jung HH, Ahn S, Bae S, Lee SK, Kim SW, Lee JE, Nam SJ, Ahn JS, Im YH, Park YH (2016) The relationship between nuclear factor (NF)-kappaB family gene expression and prognosis in triple-negative breast cancer (TNBC) patients receiving adjuvant doxorubicin treatment. Sci Rep 6:31804. https://doi.org/10.1038/srep31804
Vollebergh MA, Lips EH, Nederlof PM, Wessels LF, Schmidt MK, van Beers EH, Cornelissen S, Holtkamp M, Froklage FE, de Vries EG, Schrama JG, Wesseling J, van de Vijver MJ, van Tinteren H, de Bruin M, Hauptmann M, Rodenhuis S, Linn SC (2011) An aCGH classifier derived from BRCA1-mutated breast cancer and benefit of high-dose platinum-based chemotherapy in HER2-negative breast cancer patients. Ann Oncol 22:1561–1570. https://doi.org/10.1093/annonc/mdq624
Park YH, Im SA, Kim SB, Sohn JH, Lee KS, Chae YS, Lee KH, Kim JH, Im YH, Kim JY, Kim TY, Lee KH, Ahn JH, Kim GM, Park IH, Lee SJ, Han HS, Kim SH, Jung KH, Korean Cancer Study G (2017) Phase II, multicentre, randomised trial of eribulin plus gemcitabine versus paclitaxel plus gemcitabine as first-line chemotherapy in patients with HER2-negative metastatic breast cancer. Eur J Cancer 86:385–393. https://doi.org/10.1016/j.ejca.2017.10.002
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164. https://doi.org/10.1093/nar/gkq603
Hanzelmann S, Castelo R, Guinney J (2013) GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform 14:7. https://doi.org/10.1186/1471-2105-14-7
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG, Exome Aggregation C (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536:285–291. https://doi.org/10.1038/nature19057
Bamford S, Dawson E, Forbes S, Clements J, Pettett R, Dogan A, Flanagan A, Teague J, Futreal PA, Stratton MR, Wooster R (2004) The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer 91:355–358. https://doi.org/10.1038/sj.bjc.6601894
Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jager N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdes-Mas R, Buuren MM, Veer L, Vincent-Salomon A, Waddell N, Yates LR, Australian Pancreatic Cancer Genome I, Consortium IBC, Consortium IM-S, PedBrain I, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR (2013) Signatures of mutational processes in human cancer. Nature 500:415–421. https://doi.org/10.1038/nature12477
Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C (2016) DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol 17:31. https://doi.org/10.1186/s13059-016-0893-4
Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, Menzies A, Martin S, Leung K, Chen L, Leroy C, Ramakrishna M, Rance R, Lau KW, Mudie LJ, Varela I, McBride DJ, Bignell GR, Cooke SL, Shlien A, Gamble J, Whitmore I, Maddison M, Tarpey PS, Davies HR, Papaemmanuil E, Stephens PJ, McLaren S, Butler AP, Teague JW, Jonsson G, Garber JE, Silver D, Miron P, Fatima A, Boyault S, Langerod A, Tutt A, Martens JW, Aparicio SA, Borg A, Salomon AV, Thomas G, Borresen-Dale AL, Richardson AL, Neuberger MS, Futreal PA, Campbell PJ, Stratton MR, Breast Cancer Working Group of the International Cancer Genome C (2012) Mutational processes molding the genomes of 21 breast cancers. Cell 149:979–993. https://doi.org/10.1016/j.cell.2012.04.024
Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, Martincorena I, Alexandrov LB, Martin S, Wedge DC, Van Loo P, Ju YS, Smid M, Brinkman AB, Morganella S, Aure MR, Lingjaerde OC, Langerod A, Ringner M, Ahn SM, Boyault S, Brock JE, Broeks A, Butler A, Desmedt C, Dirix L, Dronov S, Fatima A, Foekens JA, Gerstung M, Hooijer GK, Jang SJ, Jones DR, Kim HY, King TA, Krishnamurthy S, Lee HJ, Lee JY, Li Y, McLaren S, Menzies A, Mustonen V, O’Meara S, Pauporte I, Pivot X, Purdie CA, Raine K, Ramakrishnan K, Rodriguez-Gonzalez FG, Romieu G, Sieuwerts AM, Simpson PT, Shepherd R, Stebbings L, Stefansson OA, Teague J, Tommasi S, Treilleux I, Van den Eynden GG, Vermeulen P, Vincent-Salomon A, Yates L, Caldas C, Veer L, Tutt A, Knappskog S, Tan BK, Jonkers J, Borg A, Ueno NT, Sotiriou C, Viari A, Futreal PA, Campbell PJ, Span PN, Van Laere S, Lakhani SR, Eyfjord JE, Thompson AM, Birney E, Stunnenberg HG, van de Vijver MJ, Martens JW, Borresen-Dale AL, Richardson AL, Kong G, Thomas G, Stratton MR (2016) Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534:47–54. https://doi.org/10.1038/nature17676
Eroles P, Bosch A, Perez-Fidalgo JA, Lluch A (2012) Molecular biology in breast cancer: intrinsic subtypes and signaling pathways. Cancer Treat Rev 38:698–707. https://doi.org/10.1016/j.ctrv.2011.11.005
Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X, Chen K, Scheet P, Vattathil S, Liang H, Multani A, Zhang H, Zhao R, Michor F, Meric-Bernstam F, Navin NE (2014) Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature 512:155–160. https://doi.org/10.1038/nature13600
Yoshida T, Ozawa Y, Kimura T, Sato Y, Kuznetsov G, Xu S, Uesugi M, Agoulnik S, Taylor N, Funahashi Y, Matsui J (2014) Eribulin mesilate suppresses experimental metastasis of breast cancer cells by reversing phenotype from epithelial-mesenchymal transition (EMT) to mesenchymal-epithelial transition (MET) states. Br J Cancer 110:1497–1505. https://doi.org/10.1038/bjc.2014.80
Funding
This research was supported by a Grant from the National Research Foundation of Republic of Korea (NRF-2018R1A2B6004690), the Ministry of Health and Welfare, Republic of Korea (HA17C0055), and the National R&D Program for Cancer Control, Ministry of Health and Welfare, Republic of Korea (1720150). This study was also supported by a Grant from Eisai Korea Inc.
Author information
Authors and Affiliations
Consortia
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards our institutional and/or national research committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 3 (TIFF 1477 kb)
Supplementary Fig. 1. OS and PFS according to FAT3 mutation
Supplementary material 4 (TIFF 3252 kb)
Supplementary Fig. 2. DEG analysis for angiogenesis-related genes in immune panel according to treatment arm and disease progression
Supplementary material 5 (TIFF 2385 kb)
Supplementary Fig. 3. DEG analysis for angiogenesis-related genes in cancer panel according to treatment arm and disease progression
Supplementary material 6 (TIFF 1748 kb)
Supplementary Fig. 4. GSVA for nanostring (a) cancer panel and (b) immune panel
Supplementary material 7 (TIFF 2516 kb)
Supplementary Fig. 5. GSVA for (a) cancer panel and (b) immune panel using pre- and post-treatment samples
Supplementary material 8 (TIFF 1577 kb)
Supplementary Fig. 6. Cox-regression and KM analysis according to mutation signature 3 in (a) EG arm and (b) PG arm
Rights and permissions
About this article
Cite this article
Kim, JY., Lee, E., Park, K. et al. Exploratory biomarker analysis from a phase II clinical trial of eribulin plus gemcitabine versus paclitaxel plus gemcitabine for HER2-negative metastatic breast cancer patients (KCSG BR13-11). Breast Cancer Res Treat 178, 367–377 (2019). https://doi.org/10.1007/s10549-019-05400-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-019-05400-y