Summary
Introduction
Genes that are expressed in a highly tissue- or disease-specific manner provide possible targets for therapeutics, early detection of cancer, and monitoring of disease burden during and after treatment. Further, genes of this type that code for secreted or shed proteins may allow for serum detection of the product facilitating our ability to specifically detect the cancer in all circumstances. To this end, we are working towards identification and characterization of such genes that are specifically expressed in breast epithelium. In the current study, we have measured the expression of two markers that emerged from a screen of the Incyte LifeSeq Database and were subsequently shown to be highly restricted to breast epithelium termed BU101 (also called Lipophilin B) and BS106 (small mucin-like protein). These two novel markers were compared with two other candidate markers, Mammaglobin and Cytokeratin 19 (CK19).
Methods
Utilizing quantitative real-time PCR, we compared the expression of these four genes in a series of 95 primary breast cancers, 9 lymph nodes from breast cancer patients, 13 lymph nodes from non-cancer patients and 10 normal breast tissues.
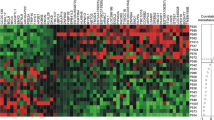
Results
Cytokeratin was shown to be highly sensitive in detecting all breast cancers, while BU101, BS106 and Mammaglobin were more restricted.
Conclusion
While no one of the these markers efficiently detects all breast cancers, a combination of two or more could achieve a very high sensitivity in assaying for circulating or occult breast cancer cells.
Similar content being viewed by others
References
Schulze R, Schulze M, Wischnik A, Ehnle S, Doukas K, Behr W, Ehret W, Schlimok G, 1997 Tumor cell contamination of peripheral blood stem cell transplants and bone marrow in high-risk breast cancer patients Bone Marrow Transpl 19: 1223–1228
Vannucchi AM, Bosi A, Glinz S, Pacini P, Linari S, Saccardi R, Alterini R, Rigacci L, Guidi S, Lombardini L, Longo G, Mariani MP, Rossi-Ferrini P, 1998: Evaluation of breast tumour cell contamination in the bone marrow and leukapheresis collections by RT-PCR for cytokeratin-19 mRNA Brit J Haematol 103: 610–617
Shammas FV, Deak E, Nysted A, van Eekelen JA, Osland A, Heikkila R, 2001 Serial quantitative PCR analysis of bone marrow samples from breast cancer patients to monitor systemic micrometastases Anticancer Res 21: 2099–2106
Ikeda N, Miyoshi Y, Motomura K, Inaji H, Koyama H, Noguchi S, 2000 Prognostic significance of occult bone marrow micrometastases of breast cancer detected by quantitative polymerase chain reaction for cytokeratin 19 mRNA Jpn J Cancer Res 91: 918–924
Kahn HJ, Yang LY, Lickley L, Holloway C, Hanna W, Narod S, McCready DR, Seth A, Marks A, 2000 RT-PCR amplification of CK19 mRNA in the blood of breast cancer patients: correlation with established prognostic parameters Breast Cancer Res Treat 60: 143–151
Colpitts TL, Billing-Medel P, Friedman P, Granados EN, Hayden M, Hodges S, Menhart N, Roberts L, Russell J, Stroupe SD, 2001 Mammaglobin is found in breast tissue as a complex with BU101 Biochem 40: 11048–11059
Zhao C, Nguyen T, Yusifov T, Glasgow BJ, Lehrer RI, 1999 Lipophilins: human peptides homologous to rat prostatein Biochem Biophys Res Commun 256: 147–155
Mukherjee AB, Kundu GC, Mantile-Selvaggi G, Yuan CJ, Mandal AK, Chattopadhyay S, Zheng F, Pattabiraman N, Zhang Z, 1999 Uteroglobin: a novel cytokine? Cell Mol Life Sci 55: 771–787
Mukherjee AB, Kundu GC, Mandal AK, Pattabiraman N, Yuan CJ, Zhang Z, 1998 Uteroglobin: physiological role in normal glomerular function uncovered by targeted disruption of the uteroglobin gene in mice Am J Kidney Dis 32: 1106–1120
Miele L, 2000 Antiflammins. Bioactive peptides derived from uteroglobin Ann N Y Acad Sci 923: 128–140
Miele L, Cordella-Miele E, Mukherjee AB, 1987 Uteroglobin: structure, molecular biology, and new perspectives on its function as a phospholipase A2 inhibitor Endocr Rev 8: 474–490
Vasanthakumar G, Manjunath R, Mukherjee AB, Warabi H, Schiffmann E, 1988 Inhibition of phagocyte chemotaxis by uteroglobin, an inhibitor of blastocyst rejection Biochem Pharmacol 37: 389–394
Levin SW, Butler JD, Schumacher UK, Wightman PD, Mukherjee AB, 1986 Uteroglobin inhibits phospholipase A2 activity Life Sci 38: 1813–1819
Watson MA, Fleming TP, 1996 Mammaglobin, a mammary-specific member of the uteroglobin gene family, is overexpressed in human breast cancer Cancer Res 56: 860–865
Colpitts TL, Billing P, Granados E, Hayden M, Hodges S, Roberts L, Russell J, Friedman P, Stroupe S, 2002 Identification and immunohistochemical characterization of a mucin-like glycoprotein expressed in early stage breast carcinoma Tumor Biol 23: 263–278
Miksicek RJ, Myal Y, Watson PH, Walker C, Murphy LC, Leygue E, 2002 Identification of a novel breast- and salivary gland-specific, mucin- like gene strongly expressed in normal and tumor human mammary epithelium Cancer Res 62: 2736–2740
Iglehart JD, Kerns BJ, Huper G, Marks JR, 1995 Maintenance of DNA content and erbB-2 alterations in intraductal and invasive phases of mammary cancer Breast Cancer Res Treat 34: 253–263
Davidoff AM, Humphrey PA, Iglehart JD, Marks JR, 1991 Genetic basis for p53 overexpression in human breast cancer Proc Natl Acad Sci U S A 88: 5006–5010
Davidoff AM, Kerns BJ, Iglehart JD, Marks JR, 1991 Maintenance of p53 alterations throughout stages of breast cancer progression Cancer Res 51: 2605–2610
Braun S, Pantel K, Muller P, Janni W, Hepp F, Kentenich CR, Gastroph S, Wischnik A, Dimpfl T, Kindermann G, Riethmuller G, Schlimok G, 2000 Cytokeratin-positive cells in the bone marrow and survival of patients with stage I, II, or III breast cancer N Engl J Med 342: 525–533
Schreiber RH, Pendas S, Ku NN, Reintgen DS, Shons AR, Berman C, Boulware D, Cox CE, 1999 Microstaging of breast cancer patients using cytokeratin staining of the sentinel lymph node. [see comments] Ann Surg Oncol 6: 95–101
Colpaert C, Vermeulen P, Jeuris W, van Beest P, Goovaerts G, Weyler J, Van Dam P, Dirix L, Van Marck E, 2001 Early distant relapse in “node-negative” breast cancer patients is not predicted by occult axillary lymph node metastases, but by the features of the primary tumour J Pathol 193: 442–449
Lambrechts AC, Bosma AJ, Klaver SG, Top B, Perebolte L, van’t Veer LJ, Rodenhuis S, 1999 Comparison of immunocytochemistry, RT-PCR, and nucleic acid sequence-based amplification for the detection of circulating breast cancer cells Breast Cancer Res Treat 56: 219–231
Grunewald K, Haun M, Urbanek M, Fiegl M, Muller-Holzner E, Gunsilius E, Dunser M, Marth C, Gastl G, 2000 Mammaglobin gene expression: a superior marker of breast cancer cells in peripheral blood in comparison to epidermal-growth- factor receptor and cytokeratin-19 Lab Invest 80: 1071–1077
Aerts J, Wynendaele W, Paridaens R, Christiaens MR, van den Bogaert W, van Oosterom AT, Vandekerckhove F, 2001 A real-time quantitative reverse transcriptase polymerase chain reaction (RT-PCR) to detect breast carcinoma cells in peripheral blood Ann Oncol 12: 39–46
Slade MJ, Smith BM, Sinnett HD, Cross NC, Coombes RC, 1999 Quantitative polymerase chain reaction for the detection of micrometastases in patients with breast cancer J Clin Oncol 17: 870–879
Acknowledgements
The authors would like to thank Nancy Glover for her excellent technical assistance. This work was supported by an Early Detection Research Network grant to J.R.M., NIH UO1 CA84955.
Author information
Authors and Affiliations
Corresponding author
Additional information
These two authors contributed equally to the work
Rights and permissions
About this article
Cite this article
Brown, N.M., Stenzel, T.T., Friedman, P.N. et al. Evaluation of expression based markers for the detection of breast cancer cells. Breast Cancer Res Treat 97, 41–47 (2006). https://doi.org/10.1007/s10549-005-9085-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-005-9085-8