Abstract
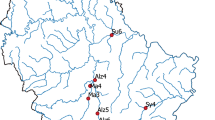
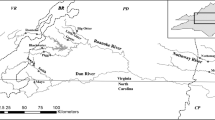
The sensitivity and specificity of eDNA-based monitoring, coupled with its potential utility to estimate population density or biomass, makes it a useful tool in invasive species management. In this study, we investigated the potential of the eDNA method to improve the detection of the elusive invasive fish, oriental weatherloach (Misgurnus anguillicaudatus), in a river system where a density gradient of the species occurs. We compared detection rates between eDNA and conventional monitoring methods and examined the relationship between eDNA and abundance in a flowing environment. The eDNA method had a higher site detection rate than conventional methods (63 vs. 38%). Weatherloach eDNA was detected at all sites where the fish has been previously caught and none of the sites where the species has not been caught for the past 7 years. There was an increasing density trend going downstream based on long-term conventional monitoring, but the eDNA concentration in water samples reflected this trend only in a continuous section of the river where impoundments were absent. We did not find a positive relationship between eDNA concentration and contemporary abundance estimates in our study area. A high eDNA concentration was recorded at a site (DVC) which was designated a low density site based on long-term catch data. This discrepancy was a likely result of physical habitat characteristics which influenced the efficiency of the conventional methods used. This study highlighted the challenges of inferring density from eDNA data in flowing water because habitat features may confound results, necessitating careful consideration for results to be useful to management.





Similar content being viewed by others
References
Abilhoa V, Bornatowski H, Vitule J (2012) Occurrence of the alien invasive loach Misgurnus anguillicaudatus in the Iguacu River basin in southern Brazil: a note of concern. J Appl Ichthyol 1:257–259. https://doi.org/10.1111/jai.12007
Balasingham KD, Walter RP, Heath DD (2017) Residual eDNA detection sensitivity assessed by quantitative real-time PCR in a river ecosystem. Mol Ecol Resour 17:523–532
Baldigo BP, Sporn LA, George SD, Ball JA (2017) Efficacy of environmental DNA to detect and quantify brook trout populations in headwater streams of the Adirondack Mountains, New York. Trans Am Fish Soc 146:99–111
Bohmann K, Evans A, Gilbert MTP, Carvalho GR, Creer S, Knapp M, Yu DW, de Bruyn M (2014) Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol Evol 29:358–367
Breen MJ, Ruetz CR III (2006) Gear bias in fyke netting: evaluating soak time, fish density, and predators. North Am J Fish Manag 26:32–41
Broadhurst B, Clear R, Fulton C, Lintermans M (2016) Enlarged cotter reservoir ecological monitoring program: technical report 2016. University of Canberra, Canberra
Bylemans J, Furlan EM, Pearce L, Daly T, Gleeson DM (2016) Improving the containment of a freshwater invader using environmental DNA (eDNA) based monitoring. Biol Invasions 18:3081–3089
Darling JA, Blum MJ (2007) DNA-based methods for monitoring invasive species: a review and prospectus. Biol Invasions 9:751–765
Deiner K, Fronhofer EA, Mächler E, Walser J-C, Altermatt F (2016) Environmental DNA reveals that rivers are conveyer belts of biodiversity information. Nat Commun 7:12544
Doi H, Inui R, Akamatsu Y, Kanno K, Yamanaka H, Takahara T, Minamoto T (2017) Environmental DNA analysis for estimating the abundance and biomass of stream fish. Freshw Biol 62:30–39
Fredberg J, Thwaites L, Earl J (2014) Oriental weatherloach, Misgurnus anguillicaudatus, in the river Murray, South Australia: a risk assessment. SARDI research report series-South Australian Research and Development Institute
Froese R, Pauly D (2016) Misgurnus anguillicaudatus. Fishbase. http://www.fishbase.org/summary/3016. Accessed 22 Dec 2016
Fujimoto Y, Ouchi Y, Hakuba T, Chiba H, Iwata M (2008) Influence of modern irrigation, drainage system and water management on spawning migration of mud loach, Misgurnus anguillicaudatus. Environ Biol Fish 81:185–194
Furlan E, Gleeson D (2016) Improving reliability in environmental DNA detection surveys through enhanced quality control. Mar Freshw Res 68:388–395. https://doi.org/10.1071/MF15349
Furlan EM, Gleeson D, Hardy CM, Duncan RP (2015) A framework for estimating the sensitivity of eDNA surveys. Mol Ecol Resour 16:641–654
Goldberg CS, Pilliod DS, Arkle RS, Waits LP (2011) Molecular detection of vertebrates in stream water: a demonstration using Rocky Mountain Tailed Frogs and Idaho Giant Salamanders. PLoS ONE. https://doi.org/10.1371/journal.pone.0022746
Goldberg CS, Strickler KM, Pilliod DS (2015) Moving environmental DNA methods from concept to practice for monitoring aquatic macroorganisms. Biol Conserv 183:1–3. https://doi.org/10.1016/j.biocon.2014.11.040
Gwinn DC, Beesley LS, Close P, Gawne B, Davies PM (2016) Imperfect detection and the determination of environmental flows for fish: challenges, implications and solutions. Freshw Biol 61:172–180
Hinlo R, Furlan E, Suitor L, Gleeson D (2017) Environmental DNA monitoring and management of invasive fish: comparison of eDNA and fyke netting. Manag Biol Invasions 8:89–100. https://doi.org/10.3391/mbi.2017.8.1.09
Hinlo R, Gleeson D, Lintermans M, Furlan E (2018) eDNA production, persistence and low-density detection of an elusive fish in an upland stream. Manuscript in preparation
ISSG (2005) Global invasive species database. Invasive Species Specialist Group (ISSG) of the IUCN Species Survival Commission. http://www.iucngisd.org/gisd/. Accessed 5 Jan 2017
Jane SF, Wilcox TM, McKelvey KS, Young MK, Schwartz MK, Lowe WH, Letcher BH, Whiteley AR (2015) Distance, flow and PCR inhibition: eDNA dynamics in two headwater streams. Mol Ecol Resour 15:216–227
Jerde CL, Mahon AR (2015) Improving confidence in environmental DNA species detection. Mol Ecol Resour 15:461–463
Jerde CL, Mahon AR, Chadderton WL, Lodge DM (2011) “Sight-unseen” detection of rare aquatic species using environmental DNA. Conserv Lett 4:150–157. https://doi.org/10.1111/j.1755-263X.2010.00158.x
Jerde CL, Chadderton WL, Mahon AR, Renshaw MA, Corush J, Budny ML, Mysorekar S, Lodge DM (2013) Detection of Asian carp DNA as part of a Great Lakes basin-wide surveillance program. Can J Fish Aquat Sci 70:522–526. https://doi.org/10.1139/cjfas-2012-0478
Keller R, Lake P (2007) Potential impacts of a recent and rapidly spreading coloniser of Australian freshwaters: oriental weatherloach (Misgurnus anguillicaudatus). Ecol Freshw Fish 16:124–132
Lacoursière-Roussel A, Côté G, Leclerc V, Bernatchez L (2015) Quantifying relative fish abundance with eDNA: a promising tool for fisheries management. J Appl Ecol 53:1148–1157
Laramie MB, Pilliod DS, Goldberg CS (2015) Characterizing the distribution of an endangered salmonid using environmental DNA analysis. Biol Conserv 183:29–37. https://doi.org/10.1016/j.biocon.2014.11.025
Leuven RS, van der Velde G, Baijens I, Snijders J, van der Zwart C, Lenders HR, bij de Vaate A (2009) The river Rhine: a global highway for dispersal of aquatic invasive species. Biol Invasions 11:1989
Lintermans M (1993) Oriental weatherloach Misgurnus anguillicaudatus in the Cotter River: A new population in the Canberra region. Technical report 4. ACT Parks and Conservation Service, Canberra, ACT, Australia
Lintermans M (2000a) Macquarie perch survey of the Paddys River. Unpublished internal report. ACT Parks and Conservation Service, Canberra, ACT, Australia
Lintermans M (2000b) The status of fish in the Australian Capital Territory: a review of current knowledge and management requirements. Technical report 15. Environment ACT, Lyneham, ACT, Australia
Lintermans M (2004) Human-assisted dispersal of alien freshwater fish in Australia. NZ J Mar Freshwat Res 38:481–501
Lintermans M (2007) Fishes of the Murray–Darling Basin: an introductory guide. Murray–Darling Basin Commission, Canberra, ACT, Australia
Lintermans M (2016) Finding the needle in the haystack: comparing sampling methods for detecting an endangered freshwater fish. Mar Freshw Res 67:1740–1749
Lintermans M, Rutzou T, Kukolic K (1990) The status, distribution and possible impacts of the oriental weatherloach Misgurnus anguillicaudatus in the Ginninderra creek catchment. Research report 2. Research Report 2. ACT Parks and Conservation Service, Canberra
Lintermans M, Raadik T, Morgan D, Jackson P (2007) Overview of the ecology and impact of three alien fish species: redfin perch, Mozambique mouthbrooder (Tilapia) and oriental weatherloach. In: Ansell D, Jackson P (eds) Emerging issues in alien fish management in the Murray–Darling Basin: statement, recommendations and supporting papers. Murray–Darling Basin Commission, Canberra, pp 22–32
Lovell SJ, Stone SF, Fernandez L (2006) The economic impacts of aquatic invasive species: a review of the literature. Agric Resour Econ Rev 35:195–208
Lyon JP, Bird T, Nicol S, Kearns J, O’Mahony J, Todd CR, Cowx IG, Bradshaw CJ (2014) Efficiency of electrofishing in turbid lowland rivers: implications for measuring temporal change in fish populations. Can J Fish Aquat Sci 71:878–886
Maruyama A, Nakamura K, Yamanaka H, Kondoh M, Minamoto T (2014) The release rate of environmental DNA from juvenile and adult fish. PLoS ONE 9:e114639
Mehta SV, Haight RG, Homans FR, Polasky S, Venette RC (2007) Optimal detection and control strategies for invasive species management. Ecol Econ 61:237–245
Meyer L, Hinrichs D (2000) Microhabitat preferences and movements of the weatherfish, Misgurnus fossilis, in a drainage channel. Environ Biol Fishes 58:297–306
Miranda L, Dolan C (2003) Test of a power transfer model for standardized electrofishing. Trans Am Fish Soc 132:1179–1185
Naruse M, Oishi T (1996) Annual and daily activity rhythms of loathes in an irrigation creek and ditches around paddy fields. Environ Biol Fish 47:93–99
Nevers MB, Byappanahalli MN, Morris CC, Shively D, Przybyla-Kelly K, Spoljaric AM, Dickey J, Roseman EF (2018) Environmental DNA (eDNA): a tool for quantifying the abundant but elusive round goby (Neogobius melanostomus). PLoS ONE 13:e0191720
Nichols S, Norris R, Maher W, Thoms M (2006) Ecological effects of serial impoundment on the Cotter River, Australia. Hydrobiologia 572:255–273
Olson ZH, Briggler JT, Williams RN (2012) An eDNA approach to detect eastern hellbenders (Cryptobranchus a. alleganiensis) using samples of water. Wildl Res 39:629–636. https://doi.org/10.1071/wr12114
Paller MH (1995) Relationships among number of fish species sampled, reach length surveyed, and sampling effort in South Carolina coastal plain streams. North Am J Fish Manag 15:110–120
Pilliod DS, Goldberg CS, Arkle RS, Waits LP (2013) Estimating occupancy and abundance of stream amphibians using environmental DNA from filtered water samples. Can J Fish Aquat Sci 70:1123–1130. https://doi.org/10.1139/cjfas-2013-0047
Pilliod DS, Goldberg CS, Arkle RS, Waits LP (2014) Factors influencing detection of eDNA from a stream-dwelling amphibian. Mol Ecol Resour 14:109–116. https://doi.org/10.1111/1755-0998.12159
Pimentel D, Lach L, Zuniga R, Morrison D (2000) Environmental and economic costs of nonindigenous species in the United States. Bioscience 50:53–65
Portt C, Coker G, Ming D, Randall R (2006) A review of fish sampling methods commonly used in Canadian freshwater habitats. Canadian technical report of fisheries and aquatic sciences 2604
Rees HC, Bishop K, Middleditch DJ, Patmore JR, Maddison BC, Gough KC (2014) The application of eDNA for monitoring of the Great Crested Newt in the UK. Ecol Evol 4:4023–4032
Roussel JM, Paillisson JM, Treguier A, Petit E (2015) The downside of eDNA as a survey tool in water bodies. J Appl Ecol 52:823–826
Sigsgaard EE, Carl H, Møller PR, Thomsen PF (2015) Monitoring the near-extinct European weather loach in Denmark based on environmental DNA from water samples. Biol Conserv 183:46–52. https://doi.org/10.1016/j.biocon.2014.11.023
Smith JM, Wells SP, Mather ME, Muth RM (2014) Fish biodiversity sampling in stream ecosystems: a process for evaluating the appropriate types and amount of gear. Aquat Conserv Mar Freshw Ecosyst 24:338–350
Spear SF, Groves JD, Williams LA, Waits LP (2015) Using environmental DNA methods to improve detectability in a hellbender (Cryptobranchus alleganiensis) monitoring program. Biol Conserv 183:38–45. https://doi.org/10.1016/j.biocon.2014.11.016
Stoner A (2004) Effects of environmental variables on fish feeding ecology: implications for the performance of baited fishing gear and stock assessment. J Fish Biol 65:1445–1471
Takahara T, Minamoto T, Yamanaka H, Doi H, Zi Kawabata (2012) Estimation of fish biomass using environmental DNA. PLoS ONE. https://doi.org/10.1371/journal.pone.0035868
Thomsen PF, Kielgast J, Iversen LL, Wiuf C, Rasmussen M, Gilbert MTP, Orlando L, Willerslev E (2012) Monitoring endangered freshwater biodiversity using environmental DNA. Mol Ecol 21:2565–2573. https://doi.org/10.1111/j.1365-294X.2011.05418.x
Turner CR, Lodge DM, Xu C, Cooper MJ, Lamberti GA (2012) Evaluating environmental DNA detection alongside standard fish sampling in Great Lakes coastal wetland monitoring (seed project). http://iiseagrant.org/research/reports/Lodge2012_finalreport_eDNA.pdf. Accessed 5 Oct 2016
Urquhart AN (2013) Life history and environmental tolerance of the invasive Oriental Weatherfish (Misgurnus anguillicaudatus) in Southwestern Idaho, USA. Boise State University, Boise
Urquhart AN, Koetsier P (2014) Low-temperature tolerance and critical thermal minimum of the invasive oriental weatherfish Misgurnus anguillicaudatus in Idaho, USA. Trans Am Fish Soc 143:68–76
Wilcox TM, McKelvey KS, Young MK, Sepulveda AJ, Shepard BB, Jane SF, Whiteley AR, Lowe WH, Schwartz MK (2016) Understanding environmental DNA detection probabilities: a case study using a stream-dwelling char Salvelinus fontinalis. Biol Conserv 194:209–216
Acknowledgements
This project was funded by the Invasive Animals Cooperative Research Centre (Project 1.W.2) and the Holsworth Wildlife Research Endowment. Conventional survey of oriental weatherloach was undertaken as part of a monitoring program funded by Icon Water. The authors are grateful to Ugyen Lhendup, Rhian Clear, Hugh Allan, Allan Couch, and Vernon Arguelles for their field assistance and Richard Duncan for his help with data analysis. Authorization to conduct the research in the Cotter and Paddys River were granted through ACT Government Scientific License Nos. FS20151, LT2015843, LT2008287; LT2010411; LT2011505; LT2013708; LT2014789; and LT2015843.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hinlo, R., Lintermans, M., Gleeson, D. et al. Performance of eDNA assays to detect and quantify an elusive benthic fish in upland streams. Biol Invasions 20, 3079–3093 (2018). https://doi.org/10.1007/s10530-018-1760-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-018-1760-x